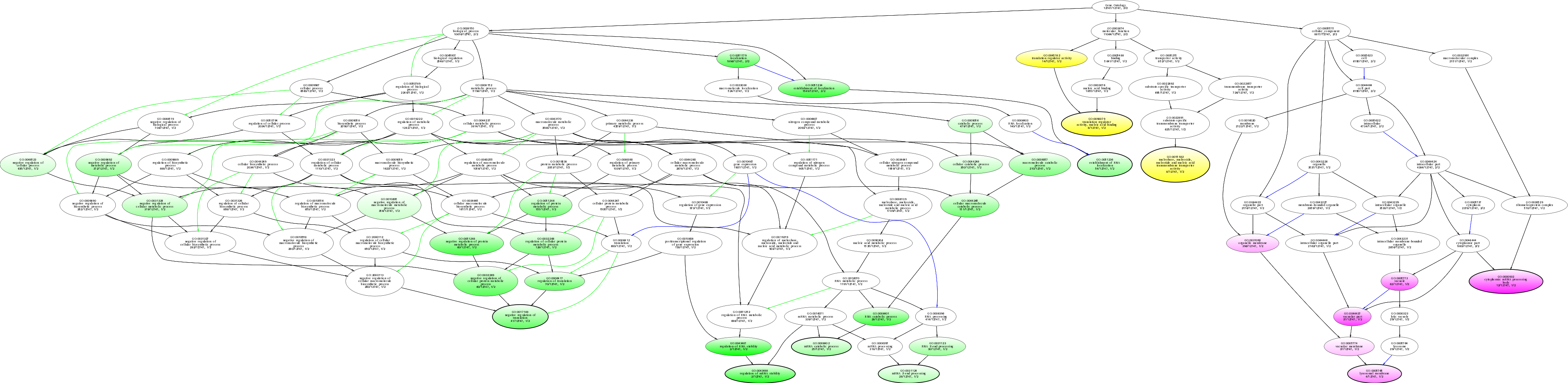
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0043487
|
regulation of RNA stability
|
0.00252845 |
0.00252845 |
1
|
2
|
|
GO:0045182
|
translation regulator activity
|
0.00252924 |
0.00252924 |
1
|
14
|
|
GO:0090079
|
translation regulator activity, nucleic acid binding
|
0.00430571 |
0.00430571 |
1
|
8
|
|
GO:0043488
|
regulation of mRNA stability
|
0.00589971 |
0.00589971 |
1
|
2
|
|
GO:0000932
|
cytoplasmic mRNA processing body
|
0.0112332 |
0.0112332 |
1
|
12
|
|
GO:0015932
|
nucleobase, nucleoside, nucleotide and nucleic acid transmembrane transporter activity
|
0.0128000 |
0.0128000 |
1
|
8
|
|
GO:0051234
|
establishment of localization
|
0.0212785 |
0.0212785 |
2
|
1549
|
|
GO:0006401
|
RNA catabolic process
|
0.0214231 |
0.0214231 |
1
|
28
|
|
GO:0044437
|
vacuolar part
|
0.0240017 |
0.0240017 |
1
|
37
|
|
GO:0051248
|
negative regulation of protein metabolic process
|
0.0257954 |
0.0257954 |
1
|
60
|
|
GO:0051179
|
localization
|
0.0318835 |
0.0318835 |
2
|
1896
|
|
GO:0005773
|
vacuole
|
0.0343417 |
0.0343417 |
1
|
62
|
|
GO:0032269
|
negative regulation of cellular protein metabolic process
|
0.0344234 |
0.0344234 |
1
|
60
|
|
GO:0006417
|
regulation of translation
|
0.0478469 |
0.0478469 |
1
|
70
|
|
GO:0017148
|
negative regulation of translation
|
0.0479532 |
0.0479532 |
1
|
41
|
|
GO:0044265
|
cellular macromolecule catabolic process
|
0.0486783 |
0.0486783 |
1
|
151
|
|
GO:0051246
|
regulation of protein metabolic process
|
0.0525602 |
0.0525602 |
1
|
155
|
|
GO:0032268
|
regulation of cellular protein metabolic process
|
0.0525883 |
0.0525883 |
1
|
128
|
|
GO:0009057
|
macromolecule catabolic process
|
0.0550603 |
0.0550603 |
1
|
210
|
|
GO:0005765
|
lysosomal membrane
|
0.0645161 |
0.0645161 |
1
|
4
|
|
GO:0051236
|
establishment of RNA localization
|
0.0657751 |
0.0657751 |
1
|
54
|
|
GO:0009892
|
negative regulation of metabolic process
|
0.0691964 |
0.0691964 |
1
|
372
|
|
GO:0031123
|
RNA 3'-end processing
|
0.0724638 |
0.0724638 |
1
|
30
|
|
GO:0006402
|
mRNA catabolic process
|
0.0730994 |
0.0730994 |
1
|
25
|
|
GO:0031324
|
negative regulation of cellular metabolic process
|
0.0793138 |
0.0793138 |
1
|
319
|
|
GO:0031090
|
organelle membrane
|
0.0800162 |
0.0800162 |
1
|
396
|
|
GO:0031124
|
mRNA 3'-end processing
|
0.0875000 |
0.0875000 |
1
|
28
|
|
GO:0044248
|
cellular catabolic process
|
0.0902527 |
0.0902527 |
1
|
350
|
|
GO:0009056
|
catabolic process
|
0.0922218 |
0.0922218 |
1
|
479
|
|
GO:0005774
|
vacuolar membrane
|
0.0934343 |
0.0934343 |
1
|
37
|
|
GO:0048523
|
negative regulation of cellular process
|
0.0966514 |
0.0966514 |
1
|
635
|
|
GO:0010605
|
negative regulation of macromolecule metabolic process
|
0.0988340 |
0.0988340 |
1
|
356
|
|
GO:0000289
|
nuclear-transcribed mRNA poly(A) tail shortening
|
0.108696 |
0.108696 |
1
|
5
|
|
GO:0003730
|
mRNA 3'-UTR binding
|
0.111675 |
0.111675 |
1
|
22
|
|
GO:0015205
|
nucleobase transmembrane transporter activity
|
0.125000 |
0.125000 |
1
|
1
|
|
GO:0048519
|
negative regulation of biological process
|
0.128590 |
0.128590 |
1
|
706
|
|
GO:0009890
|
negative regulation of biosynthetic process
|
0.129715 |
0.129715 |
1
|
282
|
|
GO:0010608
|
posttranscriptional regulation of gene expression
|
0.133607 |
0.133607 |
1
|
130
|
|
GO:0031327
|
negative regulation of cellular biosynthetic process
|
0.136166 |
0.136166 |
1
|
282
|
|
GO:0005215
|
transporter activity
|
0.148089 |
0.148089 |
1
|
852
|
|
GO:0010558
|
negative regulation of macromolecule biosynthetic process
|
0.165094 |
0.165094 |
1
|
280
|
|
GO:2000113
|
negative regulation of cellular macromolecule biosynthetic process
|
0.172946 |
0.172946 |
1
|
280
|
|
GO:0044444
|
cytoplasmic part
|
0.180530 |
0.180530 |
2
|
1863
|
|
GO:0019222
|
regulation of metabolic process
|
0.210401 |
0.210401 |
1
|
1242
|
|
GO:0030529
|
ribonucleoprotein complex
|
0.210589 |
0.210589 |
1
|
510
|
|
GO:0006412
|
translation
|
0.214209 |
0.214209 |
1
|
600
|
|
GO:0006396
|
RNA processing
|
0.214286 |
0.214286 |
1
|
414
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.237215 |
0.237215 |
1
|
1039
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.244709 |
0.244709 |
1
|
1110
|
|
GO:0006403
|
RNA localization
|
0.267176 |
0.267176 |
1
|
140
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.283034 |
0.283034 |
1
|
1056
|
|
GO:0016071
|
mRNA metabolic process
|
0.284635 |
0.284635 |
1
|
339
|
|
GO:0005737
|
cytoplasm
|
0.294170 |
0.294170 |
2
|
2378
|
|
GO:0003729
|
mRNA binding
|
0.302147 |
0.302147 |
1
|
197
|
|
GO:0000288
|
nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay
|
0.304348 |
0.304348 |
1
|
7
|
|
GO:0005515
|
protein binding
|
0.305974 |
0.305974 |
1
|
1726
|
|
GO:0050794
|
regulation of cellular process
|
0.306371 |
0.306371 |
1
|
2034
|
|
GO:0003676
|
nucleic acid binding
|
0.328842 |
0.328842 |
1
|
1855
|
|
GO:0003723
|
RNA binding
|
0.351482 |
0.351482 |
1
|
652
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.366791 |
0.366791 |
1
|
888
|
|
GO:0050789
|
regulation of biological process
|
0.378390 |
0.378390 |
1
|
2246
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.381695 |
0.381695 |
1
|
905
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.396252 |
0.396252 |
1
|
888
|
|
GO:0044425
|
membrane part
|
0.400435 |
0.400435 |
1
|
1398
|
|
GO:0009058
|
biosynthetic process
|
0.402387 |
0.402387 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.402965 |
0.402965 |
1
|
2093
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.405649 |
0.405649 |
1
|
1623
|
|
GO:0016070
|
RNA metabolic process
|
0.413829 |
0.413829 |
1
|
1191
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.415756 |
0.415756 |
1
|
1789
|
|
GO:0065007
|
biological regulation
|
0.422440 |
0.422440 |
1
|
2548
|
|
GO:0032991
|
macromolecular complex
|
0.428425 |
0.428425 |
1
|
2151
|
|
GO:0044422
|
organelle part
|
0.432705 |
0.432705 |
1
|
2176
|
|
GO:0044267
|
cellular protein metabolic process
|
0.440058 |
0.440058 |
1
|
1505
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.440308 |
0.440308 |
1
|
686
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.451726 |
0.451726 |
1
|
903
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.456989 |
0.456989 |
1
|
850
|
|
GO:0000323
|
lytic vacuole
|
0.467742 |
0.467742 |
1
|
29
|
|
GO:0019538
|
protein metabolic process
|
0.475452 |
0.475452 |
1
|
2053
|
|
GO:0033036
|
macromolecule localization
|
0.476467 |
0.476467 |
1
|
524
|
|
GO:0005575
|
cellular_component
|
0.478421 |
0.478421 |
2
|
8817
|
|
GO:0010468
|
regulation of gene expression
|
0.489930 |
0.489930 |
1
|
973
|
|
GO:0044237
|
cellular metabolic process
|
0.490989 |
0.490989 |
1
|
3814
|
|
GO:0005623
|
cell
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.496927 |
0.496927 |
1
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.498346 |
0.498346 |
1
|
1959
|
|
GO:0044424
|
intracellular part
|
0.500760 |
0.500760 |
2
|
4384
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.513595 |
0.513595 |
1
|
850
|
|
GO:0044249
|
cellular biosynthetic process
|
0.526807 |
0.526807 |
1
|
2034
|
|
GO:0010467
|
gene expression
|
0.531494 |
0.531494 |
1
|
1907
|
|
GO:0016020
|
membrane
|
0.567775 |
0.567775 |
1
|
2122
|
|
GO:0030371
|
translation repressor activity
|
0.571429 |
0.571429 |
1
|
8
|
|
GO:0005622
|
intracellular
|
0.583919 |
0.583919 |
2
|
4734
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.639129 |
0.639129 |
1
|
2878
|
|
GO:0043226
|
organelle
|
0.641414 |
0.641414 |
1
|
3537
|
|
GO:0043170
|
macromolecule metabolic process
|
0.690797 |
0.690797 |
1
|
3588
|
|
GO:0008150
|
biological_process
|
0.693584 |
0.693584 |
2
|
10616
|
|
GO:0031224
|
intrinsic to membrane
|
0.725322 |
0.725322 |
1
|
1014
|
|
GO:0006397
|
mRNA processing
|
0.734411 |
0.734411 |
1
|
318
|
|
GO:0008152
|
metabolic process
|
0.739170 |
0.739170 |
1
|
5194
|
|
GO:0044446
|
intracellular organelle part
|
0.743053 |
0.743053 |
1
|
2162
|
|
GO:0003674
|
molecular_function
|
0.753361 |
0.753361 |
2
|
11064
|
|
GO:0005488
|
binding
|
0.759777 |
0.759777 |
1
|
5641
|
|
GO:0000900
|
translation repressor activity, nucleic acid binding
|
0.777778 |
0.777778 |
1
|
7
|
|
GO:0022891
|
substrate-specific transmembrane transporter activity
|
0.788146 |
0.788146 |
1
|
625
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0022892
|
substrate-specific transporter activity
|
0.815728 |
0.815728 |
1
|
695
|
|
GO:0044238
|
primary metabolic process
|
0.819985 |
0.819985 |
1
|
4259
|
|
GO:0022857
|
transmembrane transporter activity
|
0.852113 |
0.852113 |
1
|
726
|
|
GO:0009987
|
cellular process
|
0.854049 |
0.854049 |
1
|
6560
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0000956
|
nuclear-transcribed mRNA catabolic process
|
0.920000 |
0.920000 |
1
|
23
|
|
GO:0043229
|
intracellular organelle
|
0.961659 |
0.961659 |
1
|
3530
|
|
GO:0016021
|
integral to membrane
|
0.984221 |
0.984221 |
1
|
998
|
|
GO:0006810
|
transport
|
0.998894 |
0.998894 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
2
|
12747
|
|
GO:0005764
|
lysosome
|
1.00000 |
1.00000 |
1
|
29
|