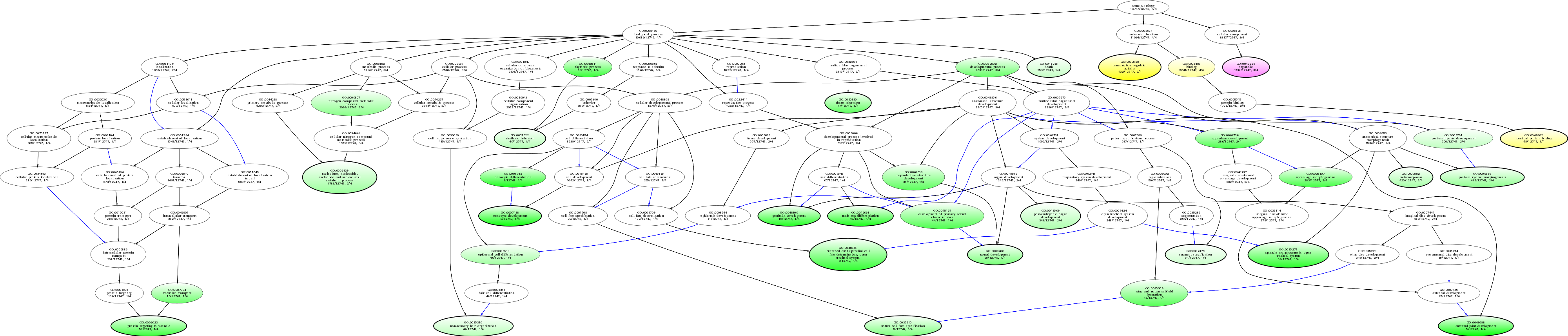
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0048098
|
antennal joint development
|
0.00661082 |
0.00661082 |
1
|
5
|
|
GO:0007438
|
oenocyte development
|
0.00767754 |
0.00767754 |
1
|
8
|
|
GO:0030528
|
transcription regulator activity
|
0.00866082 |
0.00866082 |
2
|
432
|
|
GO:0001742
|
oenocyte differentiation
|
0.0144809 |
0.0144809 |
1
|
9
|
|
GO:0046661
|
male sex differentiation
|
0.0155673 |
0.0155673 |
1
|
18
|
|
GO:0048806
|
genitalia development
|
0.0194583 |
0.0194583 |
1
|
16
|
|
GO:0035277
|
spiracle morphogenesis, open tracheal system
|
0.0223110 |
0.0223110 |
1
|
18
|
|
GO:0046845
|
branched duct epithelial cell fate determination, open tracheal system
|
0.0255682 |
0.0255682 |
1
|
9
|
|
GO:0048511
|
rhythmic process
|
0.0257497 |
0.0257497 |
1
|
69
|
|
GO:0006623
|
protein targeting to vacuole
|
0.0335570 |
0.0335570 |
1
|
5
|
|
GO:0035107
|
appendage morphogenesis
|
0.0338924 |
0.0338924 |
2
|
283
|
|
GO:0048736
|
appendage development
|
0.0340767 |
0.0340767 |
2
|
286
|
|
GO:0045137
|
development of primary sexual characteristics
|
0.0351060 |
0.0351060 |
1
|
44
|
|
GO:0035309
|
wing and notum subfield formation
|
0.0385720 |
0.0385720 |
1
|
13
|
|
GO:0048608
|
reproductive structure development
|
0.0452711 |
0.0452711 |
1
|
35
|
|
GO:0090130
|
tissue migration
|
0.0459228 |
0.0459228 |
1
|
77
|
|
GO:0032502
|
developmental process
|
0.0499048 |
0.0499048 |
3
|
2638
|
|
GO:0007034
|
vacuolar transport
|
0.0539773 |
0.0539773 |
1
|
19
|
|
GO:0042802
|
identical protein binding
|
0.0548622 |
0.0548622 |
1
|
48
|
|
GO:0008406
|
gonad development
|
0.0555886 |
0.0555886 |
1
|
35
|
|
GO:0035310
|
notum cell fate specification
|
0.0617284 |
0.0617284 |
1
|
5
|
|
GO:0043226
|
organelle
|
0.0645241 |
0.0645241 |
3
|
3537
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.0653778 |
0.0653778 |
3
|
2093
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.0671911 |
0.0671911 |
2
|
412
|
|
GO:0005488
|
binding
|
0.0675382 |
0.0675382 |
4
|
5641
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.0717945 |
0.0717945 |
3
|
1789
|
|
GO:0009913
|
epidermal cell differentiation
|
0.0726163 |
0.0726163 |
1
|
46
|
|
GO:0007552
|
metamorphosis
|
0.0748335 |
0.0748335 |
2
|
420
|
|
GO:0048569
|
post-embryonic organ development
|
0.0761074 |
0.0761074 |
2
|
343
|
|
GO:0007622
|
rhythmic behavior
|
0.0797227 |
0.0797227 |
1
|
46
|
|
GO:0035316
|
non-sensory hair organization
|
0.0907216 |
0.0907216 |
1
|
44
|
|
GO:0009791
|
post-embryonic development
|
0.0940468 |
0.0940468 |
2
|
500
|
|
GO:0016265
|
death
|
0.0940878 |
0.0940878 |
1
|
259
|
|
GO:0007379
|
segment specification
|
0.0949721 |
0.0949721 |
1
|
51
|
|
GO:0046983
|
protein dimerization activity
|
0.102695 |
0.102695 |
1
|
91
|
|
GO:0007548
|
sex differentiation
|
0.111296 |
0.111296 |
1
|
67
|
|
GO:0008219
|
cell death
|
0.112158 |
0.112158 |
1
|
255
|
|
GO:0019222
|
regulation of metabolic process
|
0.114129 |
0.114129 |
2
|
1242
|
|
GO:0006928
|
cellular component movement
|
0.118060 |
0.118060 |
1
|
269
|
|
GO:0044237
|
cellular metabolic process
|
0.118315 |
0.118315 |
3
|
3814
|
|
GO:0001708
|
cell fate specification
|
0.120037 |
0.120037 |
1
|
79
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.123669 |
0.123669 |
3
|
1959
|
|
GO:0008544
|
epidermis development
|
0.125962 |
0.125962 |
1
|
61
|
|
GO:0050954
|
sensory perception of mechanical stimulus
|
0.126761 |
0.126761 |
1
|
36
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.134439 |
0.134439 |
2
|
888
|
|
GO:0001071
|
nucleic acid binding transcription factor activity
|
0.135356 |
0.135356 |
1
|
395
|
|
GO:0008407
|
bristle morphogenesis
|
0.136320 |
0.136320 |
1
|
49
|
|
GO:0009887
|
organ morphogenesis
|
0.140035 |
0.140035 |
2
|
684
|
|
GO:0007444
|
imaginal disc development
|
0.141192 |
0.141192 |
2
|
467
|
|
GO:0045465
|
R8 cell differentiation
|
0.141791 |
0.141791 |
1
|
19
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.142051 |
0.142051 |
2
|
1039
|
|
GO:0048869
|
cellular developmental process
|
0.146194 |
0.146194 |
2
|
1276
|
|
GO:0051179
|
localization
|
0.148830 |
0.148830 |
2
|
1896
|
|
GO:0010467
|
gene expression
|
0.150029 |
0.150029 |
3
|
1907
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.150277 |
0.150277 |
2
|
1110
|
|
GO:0009888
|
tissue development
|
0.151556 |
0.151556 |
2
|
557
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.156909 |
0.156909 |
2
|
888
|
|
GO:0007469
|
antennal development
|
0.158460 |
0.158460 |
1
|
25
|
|
GO:0045475
|
locomotor rhythm
|
0.165680 |
0.165680 |
1
|
28
|
|
GO:0005681
|
spliceosomal complex
|
0.167598 |
0.167598 |
1
|
180
|
|
GO:0045184
|
establishment of protein localization
|
0.168623 |
0.168623 |
1
|
273
|
|
GO:0040011
|
locomotion
|
0.170292 |
0.170292 |
1
|
484
|
|
GO:0048563
|
post-embryonic organ morphogenesis
|
0.172051 |
0.172051 |
2
|
323
|
|
GO:0015031
|
protein transport
|
0.177333 |
0.177333 |
1
|
266
|
|
GO:0007423
|
sensory organ development
|
0.191648 |
0.191648 |
2
|
544
|
|
GO:0009892
|
negative regulation of metabolic process
|
0.193590 |
0.193590 |
1
|
372
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.194907 |
0.194907 |
2
|
1056
|
|
GO:0001709
|
cell fate determination
|
0.196268 |
0.196268 |
1
|
132
|
|
GO:0050789
|
regulation of biological process
|
0.198789 |
0.198789 |
2
|
2246
|
|
GO:0030030
|
cell projection organization
|
0.205631 |
0.205631 |
1
|
485
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.208706 |
0.208706 |
2
|
850
|
|
GO:0008586
|
imaginal disc-derived wing vein morphogenesis
|
0.215388 |
0.215388 |
1
|
47
|
|
GO:0008584
|
male gonad development
|
0.219512 |
0.219512 |
1
|
9
|
|
GO:0031324
|
negative regulation of cellular metabolic process
|
0.219618 |
0.219618 |
1
|
319
|
|
GO:0050794
|
regulation of cellular process
|
0.224039 |
0.224039 |
2
|
2034
|
|
GO:0046907
|
intracellular transport
|
0.227832 |
0.227832 |
1
|
352
|
|
GO:0022416
|
bristle development
|
0.228124 |
0.228124 |
1
|
66
|
|
GO:0048519
|
negative regulation of biological process
|
0.240665 |
0.240665 |
1
|
706
|
|
GO:0051641
|
cellular localization
|
0.242560 |
0.242560 |
1
|
607
|
|
GO:0009890
|
negative regulation of biosynthetic process
|
0.242656 |
0.242656 |
1
|
282
|
|
GO:0065007
|
biological regulation
|
0.244964 |
0.244964 |
2
|
2548
|
|
GO:0006622
|
protein targeting to lysosome
|
0.250000 |
0.250000 |
1
|
4
|
|
GO:0031327
|
negative regulation of cellular biosynthetic process
|
0.253848 |
0.253848 |
1
|
282
|
|
GO:0051674
|
localization of cell
|
0.258248 |
0.258248 |
1
|
263
|
|
GO:0035214
|
eye-antennal disc development
|
0.259257 |
0.259257 |
1
|
65
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.260978 |
0.260978 |
3
|
2878
|
|
GO:0005634
|
nucleus
|
0.261198 |
0.261198 |
3
|
1826
|
|
GO:0048523
|
negative regulation of cellular process
|
0.262869 |
0.262869 |
1
|
635
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.263629 |
0.263629 |
2
|
850
|
|
GO:0010605
|
negative regulation of macromolecule metabolic process
|
0.268230 |
0.268230 |
1
|
356
|
|
GO:0060541
|
respiratory system development
|
0.272151 |
0.272151 |
1
|
249
|
|
GO:0007626
|
locomotory behavior
|
0.280859 |
0.280859 |
1
|
157
|
|
GO:0007427
|
epithelial cell migration, open tracheal system
|
0.282258 |
0.282258 |
1
|
70
|
|
GO:0046530
|
photoreceptor cell differentiation
|
0.295620 |
0.295620 |
1
|
162
|
|
GO:0008152
|
metabolic process
|
0.296531 |
0.296531 |
3
|
5194
|
|
GO:0030529
|
ribonucleoprotein complex
|
0.298647 |
0.298647 |
1
|
510
|
|
GO:0043565
|
sequence-specific DNA binding
|
0.300971 |
0.300971 |
1
|
248
|
|
GO:0010558
|
negative regulation of macromolecule biosynthetic process
|
0.303014 |
0.303014 |
1
|
280
|
|
GO:0051649
|
establishment of localization in cell
|
0.303951 |
0.303951 |
1
|
500
|
|
GO:0051172
|
negative regulation of nitrogen compound metabolic process
|
0.306835 |
0.306835 |
1
|
250
|
|
GO:0035317
|
imaginal disc-derived wing hair organization
|
0.309372 |
0.309372 |
1
|
43
|
|
GO:0001667
|
ameboidal cell migration
|
0.310204 |
0.310204 |
1
|
76
|
|
GO:0007498
|
mesoderm development
|
0.312278 |
0.312278 |
1
|
95
|
|
GO:0045466
|
R7 cell differentiation
|
0.313433 |
0.313433 |
1
|
42
|
|
GO:2000113
|
negative regulation of cellular macromolecule biosynthetic process
|
0.316071 |
0.316071 |
1
|
280
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.325784 |
0.325784 |
2
|
905
|
|
GO:0043170
|
macromolecule metabolic process
|
0.329564 |
0.329564 |
3
|
3588
|
|
GO:0007560
|
imaginal disc morphogenesis
|
0.331061 |
0.331061 |
2
|
323
|
|
GO:0000003
|
reproduction
|
0.332995 |
0.332995 |
1
|
1022
|
|
GO:0022414
|
reproductive process
|
0.332995 |
0.332995 |
1
|
1022
|
|
GO:0046546
|
development of primary male sexual characteristics
|
0.333333 |
0.333333 |
1
|
15
|
|
GO:0006350
|
transcription
|
0.334468 |
0.334468 |
2
|
859
|
|
GO:0005623
|
cell
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0044464
|
cell part
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0045934
|
negative regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.353746 |
0.353746 |
1
|
250
|
|
GO:0044424
|
intracellular part
|
0.354324 |
0.354324 |
3
|
4384
|
|
GO:0001751
|
compound eye photoreceptor cell differentiation
|
0.354497 |
0.354497 |
1
|
134
|
|
GO:0009058
|
biosynthetic process
|
0.355414 |
0.355414 |
2
|
2090
|
|
GO:0005515
|
protein binding
|
0.358840 |
0.358840 |
2
|
1726
|
|
GO:0045165
|
cell fate commitment
|
0.359875 |
0.359875 |
1
|
255
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.360119 |
0.360119 |
2
|
1623
|
|
GO:0003008
|
system process
|
0.360531 |
0.360531 |
1
|
664
|
|
GO:0007610
|
behavior
|
0.361111 |
0.361111 |
1
|
559
|
|
GO:0001754
|
eye photoreceptor cell differentiation
|
0.361386 |
0.361386 |
1
|
146
|
|
GO:0032501
|
multicellular organismal process
|
0.369984 |
0.369984 |
2
|
3315
|
|
GO:0016070
|
RNA metabolic process
|
0.371980 |
0.371980 |
2
|
1191
|
|
GO:0003702
|
RNA polymerase II transcription factor activity
|
0.381445 |
0.381445 |
2
|
267
|
|
GO:0010629
|
negative regulation of gene expression
|
0.384189 |
0.384189 |
1
|
288
|
|
GO:0003676
|
nucleic acid binding
|
0.399422 |
0.399422 |
2
|
1855
|
|
GO:0035120
|
post-embryonic appendage morphogenesis
|
0.406614 |
0.406614 |
2
|
268
|
|
GO:0070727
|
cellular macromolecule localization
|
0.408300 |
0.408300 |
1
|
305
|
|
GO:0030539
|
male genitalia development
|
0.409091 |
0.409091 |
1
|
9
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.427778 |
0.427778 |
2
|
903
|
|
GO:0016481
|
negative regulation of transcription
|
0.428239 |
0.428239 |
1
|
233
|
|
GO:0007472
|
wing disc morphogenesis
|
0.442702 |
0.442702 |
2
|
255
|
|
GO:0005622
|
intracellular
|
0.446165 |
0.446165 |
3
|
4734
|
|
GO:0007600
|
sensory perception
|
0.448657 |
0.448657 |
1
|
284
|
|
GO:0046982
|
protein heterodimerization activity
|
0.461538 |
0.461538 |
1
|
42
|
|
GO:0035220
|
wing disc development
|
0.463216 |
0.463216 |
2
|
318
|
|
GO:0050896
|
response to stimulus
|
0.467695 |
0.467695 |
1
|
1548
|
|
GO:0051234
|
establishment of localization
|
0.467930 |
0.467930 |
1
|
1549
|
|
GO:0045449
|
regulation of transcription
|
0.472005 |
0.472005 |
2
|
771
|
|
GO:0033036
|
macromolecule localization
|
0.476467 |
0.476467 |
1
|
524
|
|
GO:0034613
|
cellular protein localization
|
0.478936 |
0.478936 |
1
|
216
|
|
GO:0008150
|
biological_process
|
0.481028 |
0.481028 |
4
|
10616
|
|
GO:0010468
|
regulation of gene expression
|
0.484889 |
0.484889 |
2
|
973
|
|
GO:0016071
|
mRNA metabolic process
|
0.488424 |
0.488424 |
1
|
339
|
|
GO:0042803
|
protein homodimerization activity
|
0.494624 |
0.494624 |
1
|
46
|
|
GO:0007389
|
pattern specification process
|
0.494958 |
0.494958 |
1
|
537
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.495389 |
0.495389 |
2
|
1617
|
|
GO:0003006
|
developmental process involved in reproduction
|
0.497485 |
0.497485 |
1
|
602
|
|
GO:0003704
|
specific RNA polymerase II transcription factor activity
|
0.499704 |
0.499704 |
1
|
78
|
|
GO:0006886
|
intracellular protein transport
|
0.504878 |
0.504878 |
1
|
207
|
|
GO:0009987
|
cellular process
|
0.506393 |
0.506393 |
3
|
6560
|
|
GO:0048870
|
cell motility
|
0.512097 |
0.512097 |
1
|
254
|
|
GO:0006396
|
RNA processing
|
0.515147 |
0.515147 |
1
|
414
|
|
GO:0043229
|
intracellular organelle
|
0.519826 |
0.519826 |
3
|
3530
|
|
GO:0043227
|
membrane-bounded organelle
|
0.528019 |
0.528019 |
3
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.528598 |
0.528598 |
3
|
2856
|
|
GO:0044249
|
cellular biosynthetic process
|
0.540182 |
0.540182 |
2
|
2034
|
|
GO:0044238
|
primary metabolic process
|
0.551267 |
0.551267 |
3
|
4259
|
|
GO:0003674
|
molecular_function
|
0.567526 |
0.567526 |
4
|
11064
|
|
GO:0032991
|
macromolecular complex
|
0.567899 |
0.567899 |
1
|
2151
|
|
GO:0048513
|
organ development
|
0.572322 |
0.572322 |
2
|
1242
|
|
GO:0044422
|
organelle part
|
0.572743 |
0.572743 |
1
|
2176
|
|
GO:0035282
|
segmentation
|
0.581028 |
0.581028 |
1
|
294
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.587518 |
0.587518 |
1
|
2108
|
|
GO:0000166
|
nucleotide binding
|
0.608293 |
0.608293 |
1
|
1178
|
|
GO:0044428
|
nuclear part
|
0.613389 |
0.613389 |
1
|
830
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.621214 |
0.621214 |
2
|
1534
|
|
GO:0048512
|
circadian behavior
|
0.623188 |
0.623188 |
1
|
43
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.631500 |
0.631500 |
3
|
1535
|
|
GO:0048856
|
anatomical structure development
|
0.632847 |
0.632847 |
3
|
2265
|
|
GO:0006915
|
apoptosis
|
0.633333 |
0.633333 |
1
|
152
|
|
GO:0005575
|
cellular_component
|
0.637024 |
0.637024 |
3
|
8817
|
|
GO:0032774
|
RNA biosynthetic process
|
0.642295 |
0.642295 |
1
|
703
|
|
GO:0006605
|
protein targeting
|
0.657005 |
0.657005 |
1
|
136
|
|
GO:0008380
|
RNA splicing
|
0.669082 |
0.669082 |
1
|
277
|
|
GO:0071013
|
catalytic step 2 spliceosome
|
0.683333 |
0.683333 |
1
|
123
|
|
GO:0030182
|
neuron differentiation
|
0.689161 |
0.689161 |
1
|
548
|
|
GO:0003677
|
DNA binding
|
0.691225 |
0.691225 |
1
|
824
|
|
GO:0022008
|
neurogenesis
|
0.700310 |
0.700310 |
1
|
632
|
|
GO:0043167
|
ion binding
|
0.709151 |
0.709151 |
1
|
1498
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.709978 |
0.709978 |
2
|
402
|
|
GO:0008104
|
protein localization
|
0.727099 |
0.727099 |
1
|
381
|
|
GO:0048731
|
system development
|
0.728680 |
0.728680 |
2
|
1696
|
|
GO:0006397
|
mRNA processing
|
0.734411 |
0.734411 |
1
|
318
|
|
GO:0007275
|
multicellular organismal development
|
0.738485 |
0.738485 |
2
|
2296
|
|
GO:0007399
|
nervous system development
|
0.750147 |
0.750147 |
1
|
848
|
|
GO:0008270
|
zinc ion binding
|
0.781440 |
0.781440 |
1
|
901
|
|
GO:0007041
|
lysosomal transport
|
0.789474 |
0.789474 |
1
|
15
|
|
GO:0048592
|
eye morphogenesis
|
0.790642 |
0.790642 |
1
|
393
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0001745
|
compound eye morphogenesis
|
0.811947 |
0.811947 |
1
|
367
|
|
GO:0007476
|
imaginal disc-derived wing morphogenesis
|
0.816751 |
0.816751 |
2
|
254
|
|
GO:0071011
|
precatalytic spliceosome
|
0.822222 |
0.822222 |
1
|
148
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.824939 |
0.824939 |
1
|
686
|
|
GO:0007605
|
sensory perception of sound
|
0.833333 |
0.833333 |
1
|
30
|
|
GO:0000398
|
nuclear mRNA splicing, via spliceosome
|
0.836478 |
0.836478 |
1
|
266
|
|
GO:0048468
|
cell development
|
0.840445 |
0.840445 |
1
|
1042
|
|
GO:0044446
|
intracellular organelle part
|
0.869797 |
0.869797 |
1
|
2162
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.900589 |
0.900589 |
1
|
620
|
|
GO:0090132
|
epithelium migration
|
0.909091 |
0.909091 |
1
|
70
|
|
GO:0002165
|
instar larval or pupal development
|
0.910028 |
0.910028 |
2
|
477
|
|
GO:0010631
|
epithelial cell migration
|
0.921053 |
0.921053 |
1
|
70
|
|
GO:0048749
|
compound eye development
|
0.932314 |
0.932314 |
1
|
427
|
|
GO:0012501
|
programmed cell death
|
0.941176 |
0.941176 |
1
|
240
|
|
GO:0003002
|
regionalization
|
0.942272 |
0.942272 |
1
|
506
|
|
GO:0030154
|
cell differentiation
|
0.942825 |
0.942825 |
2
|
1239
|
|
GO:0035114
|
imaginal disc-derived appendage morphogenesis
|
0.951564 |
0.951564 |
2
|
279
|
|
GO:0050877
|
neurological system process
|
0.953313 |
0.953313 |
1
|
633
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0035315
|
hair cell differentiation
|
0.956522 |
0.956522 |
1
|
44
|
|
GO:0000375
|
RNA splicing, via transesterification reactions
|
0.963899 |
0.963899 |
1
|
267
|
|
GO:0016477
|
cell migration
|
0.964567 |
0.964567 |
1
|
245
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.965643 |
0.965643 |
1
|
701
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0048737
|
imaginal disc-derived appendage development
|
0.972175 |
0.972175 |
2
|
282
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0001654
|
eye development
|
0.975253 |
0.975253 |
1
|
458
|
|
GO:0007623
|
circadian rhythm
|
0.985507 |
0.985507 |
1
|
68
|
|
GO:0007424
|
open tracheal system development
|
0.995984 |
0.995984 |
1
|
248
|
|
GO:0000377
|
RNA splicing, via transesterification reactions with bulged adenosine as nucleophile
|
0.996255 |
0.996255 |
1
|
266
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
4
|
12747
|
|
GO:0003700
|
sequence-specific DNA binding transcription factor activity
|
1.00000 |
1.00000 |
1
|
395
|