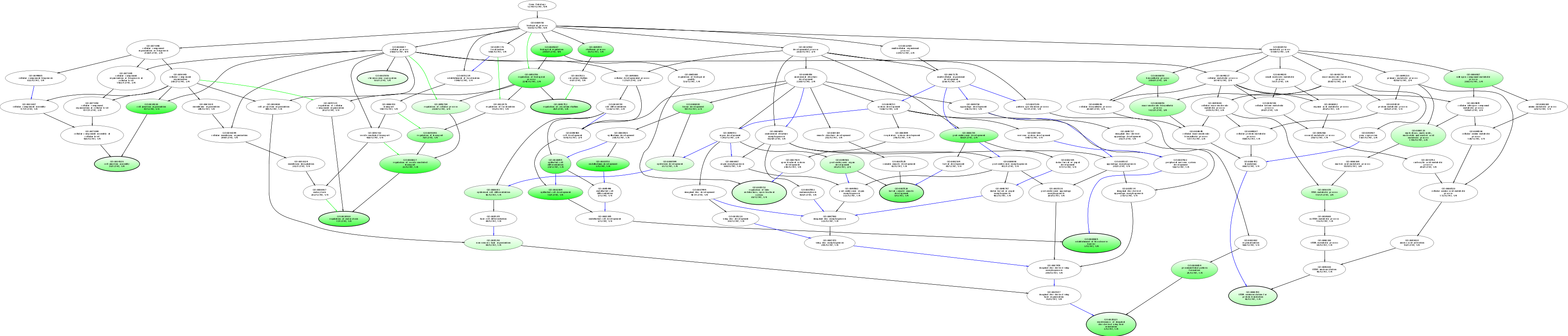
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0065007
|
biological regulation
|
0.00331266 |
0.00331266 |
4
|
2548
|
|
GO:0003158
|
endothelium development
|
0.0101695 |
0.0101695 |
1
|
3
|
|
GO:0002064
|
epithelial cell development
|
0.0190744 |
0.0190744 |
1
|
10
|
|
GO:0048511
|
rhythmic process
|
0.0257497 |
0.0257497 |
1
|
69
|
|
GO:0034330
|
cell junction organization
|
0.0283285 |
0.0283285 |
1
|
47
|
|
GO:0060627
|
regulation of vesicle-mediated transport
|
0.0295991 |
0.0295991 |
1
|
23
|
|
GO:0008065
|
establishment of blood-nerve barrier
|
0.0300000 |
0.0300000 |
1
|
3
|
|
GO:0050789
|
regulation of biological process
|
0.0318419 |
0.0318419 |
3
|
2246
|
|
GO:0030100
|
regulation of endocytosis
|
0.0322581 |
0.0322581 |
1
|
17
|
|
GO:0030855
|
epithelial cell differentiation
|
0.0350466 |
0.0350466 |
1
|
24
|
|
GO:0009791
|
post-embryonic development
|
0.0358662 |
0.0358662 |
2
|
500
|
|
GO:0007526
|
larval somatic muscle development
|
0.0385042 |
0.0385042 |
1
|
8
|
|
GO:0042752
|
regulation of circadian rhythm
|
0.0457046 |
0.0457046 |
1
|
35
|
|
GO:0035321
|
maintenance of imaginal disc-derived wing hair orientation
|
0.0476190 |
0.0476190 |
1
|
3
|
|
GO:0009954
|
proximal/distal pattern formation
|
0.0494071 |
0.0494071 |
1
|
25
|
|
GO:0051049
|
regulation of transport
|
0.0514323 |
0.0514323 |
1
|
79
|
|
GO:0009888
|
tissue development
|
0.0603928 |
0.0603928 |
2
|
557
|
|
GO:0009058
|
biosynthetic process
|
0.0650969 |
0.0650969 |
3
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.0653778 |
0.0653778 |
3
|
2093
|
|
GO:0034329
|
cell junction assembly
|
0.0660870 |
0.0660870 |
1
|
38
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.0666764 |
0.0666764 |
3
|
1623
|
|
GO:0016070
|
RNA metabolic process
|
0.0707654 |
0.0707654 |
3
|
1191
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.0717945 |
0.0717945 |
3
|
1789
|
|
GO:0009913
|
epidermal cell differentiation
|
0.0726163 |
0.0726163 |
1
|
46
|
|
GO:0048569
|
post-embryonic organ development
|
0.0761074 |
0.0761074 |
2
|
343
|
|
GO:0035152
|
regulation of tube architecture, open tracheal system
|
0.0782313 |
0.0782313 |
1
|
69
|
|
GO:0006418
|
tRNA aminoacylation for protein translation
|
0.0816667 |
0.0816667 |
1
|
49
|
|
GO:0008544
|
epidermis development
|
0.0858137 |
0.0858137 |
1
|
61
|
|
GO:0050794
|
regulation of cellular process
|
0.0885337 |
0.0885337 |
3
|
2034
|
|
GO:0035316
|
non-sensory hair organization
|
0.0907216 |
0.0907216 |
1
|
44
|
|
GO:0007059
|
chromosome segregation
|
0.0974633 |
0.0974633 |
1
|
166
|
|
GO:0016321
|
female meiosis chromosome segregation
|
0.103817 |
0.103817 |
1
|
68
|
|
GO:0016874
|
ligase activity
|
0.109366 |
0.109366 |
1
|
231
|
|
GO:0019222
|
regulation of metabolic process
|
0.114129 |
0.114129 |
2
|
1242
|
|
GO:0007422
|
peripheral nervous system development
|
0.114481 |
0.114481 |
1
|
100
|
|
GO:0032879
|
regulation of localization
|
0.114771 |
0.114771 |
1
|
143
|
|
GO:0006520
|
cellular amino acid metabolic process
|
0.115315 |
0.115315 |
1
|
172
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.122595 |
0.122595 |
3
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.123669 |
0.123669 |
3
|
1959
|
|
GO:0045446
|
endothelial cell differentiation
|
0.125000 |
0.125000 |
1
|
3
|
|
GO:0001071
|
nucleic acid binding transcription factor activity
|
0.135356 |
0.135356 |
1
|
395
|
|
GO:0016327
|
apicolateral plasma membrane
|
0.136729 |
0.136729 |
1
|
51
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.142051 |
0.142051 |
2
|
1039
|
|
GO:0009987
|
cellular process
|
0.145754 |
0.145754 |
4
|
6560
|
|
GO:0044249
|
cellular biosynthetic process
|
0.146100 |
0.146100 |
3
|
2034
|
|
GO:0048869
|
cellular developmental process
|
0.146194 |
0.146194 |
2
|
1276
|
|
GO:0030528
|
transcription regulator activity
|
0.147289 |
0.147289 |
1
|
432
|
|
GO:0010467
|
gene expression
|
0.150029 |
0.150029 |
3
|
1907
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.150277 |
0.150277 |
2
|
1110
|
|
GO:0007525
|
somatic muscle development
|
0.166667 |
0.166667 |
1
|
42
|
|
GO:0035317
|
imaginal disc-derived wing hair organization
|
0.168627 |
0.168627 |
1
|
43
|
|
GO:0043062
|
extracellular structure organization
|
0.168915 |
0.168915 |
1
|
134
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.178576 |
0.178576 |
2
|
2108
|
|
GO:0055001
|
muscle cell development
|
0.179057 |
0.179057 |
1
|
100
|
|
GO:0043226
|
organelle
|
0.180483 |
0.180483 |
3
|
3537
|
|
GO:0035107
|
appendage morphogenesis
|
0.184365 |
0.184365 |
1
|
283
|
|
GO:0006082
|
organic acid metabolic process
|
0.189003 |
0.189003 |
1
|
259
|
|
GO:0016044
|
cellular membrane organization
|
0.193867 |
0.193867 |
1
|
344
|
|
GO:0042692
|
muscle cell differentiation
|
0.194824 |
0.194824 |
1
|
132
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.194907 |
0.194907 |
2
|
1056
|
|
GO:0003677
|
DNA binding
|
0.197185 |
0.197185 |
2
|
824
|
|
GO:0042180
|
cellular ketone metabolic process
|
0.203187 |
0.203187 |
1
|
280
|
|
GO:0032774
|
RNA biosynthetic process
|
0.203435 |
0.203435 |
2
|
703
|
|
GO:0048468
|
cell development
|
0.209123 |
0.209123 |
2
|
1042
|
|
GO:0048736
|
appendage development
|
0.209532 |
0.209532 |
1
|
286
|
|
GO:0061061
|
muscle structure development
|
0.210182 |
0.210182 |
1
|
252
|
|
GO:0016564
|
transcription repressor activity
|
0.215278 |
0.215278 |
1
|
93
|
|
GO:0016192
|
vesicle-mediated transport
|
0.228410 |
0.228410 |
1
|
430
|
|
GO:0005575
|
cellular_component
|
0.228855 |
0.228855 |
4
|
8817
|
|
GO:0016875
|
ligase activity, forming carbon-oxygen bonds
|
0.229437 |
0.229437 |
1
|
53
|
|
GO:0090066
|
regulation of anatomical structure size
|
0.231824 |
0.231824 |
1
|
169
|
|
GO:0004091
|
carboxylesterase activity
|
0.233871 |
0.233871 |
1
|
116
|
|
GO:0044459
|
plasma membrane part
|
0.236675 |
0.236675 |
1
|
373
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.240586 |
0.240586 |
2
|
1562
|
|
GO:0005623
|
cell
|
0.243645 |
0.243645 |
4
|
6195
|
|
GO:0044464
|
cell part
|
0.243645 |
0.243645 |
4
|
6195
|
|
GO:0016788
|
hydrolase activity, acting on ester bonds
|
0.251521 |
0.251521 |
1
|
496
|
|
GO:0003676
|
nucleic acid binding
|
0.253252 |
0.253252 |
2
|
1855
|
|
GO:0044106
|
cellular amine metabolic process
|
0.255042 |
0.255042 |
1
|
195
|
|
GO:0032502
|
developmental process
|
0.259158 |
0.259158 |
2
|
2638
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.260978 |
0.260978 |
3
|
2878
|
|
GO:0005634
|
nucleus
|
0.261198 |
0.261198 |
3
|
1826
|
|
GO:0030030
|
cell projection organization
|
0.264325 |
0.264325 |
1
|
485
|
|
GO:0030054
|
cell junction
|
0.270777 |
0.270777 |
1
|
101
|
|
GO:0034660
|
ncRNA metabolic process
|
0.271000 |
0.271000 |
1
|
119
|
|
GO:0060541
|
respiratory system development
|
0.272151 |
0.272151 |
1
|
249
|
|
GO:0035150
|
regulation of tube size
|
0.272189 |
0.272189 |
1
|
46
|
|
GO:0007552
|
metamorphosis
|
0.273794 |
0.273794 |
1
|
420
|
|
GO:0007517
|
muscle organ development
|
0.276072 |
0.276072 |
1
|
191
|
|
GO:0051128
|
regulation of cellular component organization
|
0.289045 |
0.289045 |
1
|
282
|
|
GO:0043038
|
amino acid activation
|
0.290698 |
0.290698 |
1
|
50
|
|
GO:0005886
|
plasma membrane
|
0.292027 |
0.292027 |
1
|
641
|
|
GO:0008152
|
metabolic process
|
0.296531 |
0.296531 |
3
|
5194
|
|
GO:0044237
|
cellular metabolic process
|
0.299056 |
0.299056 |
3
|
3814
|
|
GO:0001885
|
endothelial cell development
|
0.300000 |
0.300000 |
1
|
3
|
|
GO:0060537
|
muscle tissue development
|
0.300305 |
0.300305 |
1
|
91
|
|
GO:0048513
|
organ development
|
0.300572 |
0.300572 |
2
|
1242
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.304837 |
0.304837 |
2
|
888
|
|
GO:0061024
|
membrane organization
|
0.308058 |
0.308058 |
1
|
345
|
|
GO:0002164
|
larval development
|
0.311383 |
0.311383 |
1
|
85
|
|
GO:0045132
|
meiotic chromosome segregation
|
0.315287 |
0.315287 |
1
|
99
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.325784 |
0.325784 |
2
|
905
|
|
GO:0005488
|
binding
|
0.327389 |
0.327389 |
3
|
5641
|
|
GO:0043170
|
macromolecule metabolic process
|
0.329564 |
0.329564 |
3
|
3588
|
|
GO:0006350
|
transcription
|
0.334468 |
0.334468 |
2
|
859
|
|
GO:0022402
|
cell cycle process
|
0.343522 |
0.343522 |
1
|
655
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.346544 |
0.346544 |
2
|
888
|
|
GO:0051321
|
meiotic cell cycle
|
0.357143 |
0.357143 |
1
|
270
|
|
GO:0001882
|
nucleoside binding
|
0.361738 |
0.361738 |
1
|
784
|
|
GO:0007389
|
pattern specification process
|
0.365750 |
0.365750 |
1
|
537
|
|
GO:0032501
|
multicellular organismal process
|
0.369984 |
0.369984 |
2
|
3315
|
|
GO:0044281
|
small molecule metabolic process
|
0.378352 |
0.378352 |
1
|
761
|
|
GO:0007049
|
cell cycle
|
0.387308 |
0.387308 |
1
|
756
|
|
GO:0071944
|
cell periphery
|
0.391801 |
0.391801 |
1
|
724
|
|
GO:0009308
|
amine metabolic process
|
0.397406 |
0.397406 |
1
|
325
|
|
GO:0007143
|
female meiosis
|
0.401575 |
0.401575 |
1
|
102
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.410831 |
0.410831 |
2
|
686
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.427778 |
0.427778 |
2
|
903
|
|
GO:0048731
|
system development
|
0.432376 |
0.432376 |
2
|
1696
|
|
GO:0007126
|
meiosis
|
0.432709 |
0.432709 |
1
|
254
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.435609 |
0.435609 |
2
|
850
|
|
GO:0007275
|
multicellular organismal development
|
0.442066 |
0.442066 |
2
|
2296
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.451701 |
0.451701 |
1
|
412
|
|
GO:0051327
|
M phase of meiotic cell cycle
|
0.456014 |
0.456014 |
1
|
254
|
|
GO:0051234
|
establishment of localization
|
0.467930 |
0.467930 |
1
|
1549
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.468064 |
0.468064 |
2
|
620
|
|
GO:0045449
|
regulation of transcription
|
0.472005 |
0.472005 |
2
|
771
|
|
GO:0003824
|
catalytic activity
|
0.474372 |
0.474372 |
2
|
4106
|
|
GO:0022607
|
cellular component assembly
|
0.475076 |
0.475076 |
1
|
577
|
|
GO:0070160
|
occluding junction
|
0.476190 |
0.476190 |
1
|
30
|
|
GO:0008150
|
biological_process
|
0.481028 |
0.481028 |
4
|
10616
|
|
GO:0010468
|
regulation of gene expression
|
0.484889 |
0.484889 |
2
|
973
|
|
GO:0000166
|
nucleotide binding
|
0.504833 |
0.504833 |
1
|
1178
|
|
GO:0044085
|
cellular component biogenesis
|
0.509834 |
0.509834 |
1
|
632
|
|
GO:0043565
|
sequence-specific DNA binding
|
0.511614 |
0.511614 |
1
|
248
|
|
GO:0006412
|
translation
|
0.514942 |
0.514942 |
1
|
600
|
|
GO:0043229
|
intracellular organelle
|
0.519826 |
0.519826 |
3
|
3530
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.520400 |
0.520400 |
2
|
850
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.524233 |
0.524233 |
2
|
1517
|
|
GO:0043227
|
membrane-bounded organelle
|
0.528019 |
0.528019 |
3
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.528598 |
0.528598 |
3
|
2856
|
|
GO:0060538
|
skeletal muscle organ development
|
0.539267 |
0.539267 |
1
|
103
|
|
GO:0005918
|
septate junction
|
0.543478 |
0.543478 |
1
|
25
|
|
GO:0051179
|
localization
|
0.544835 |
0.544835 |
1
|
1896
|
|
GO:0044238
|
primary metabolic process
|
0.551267 |
0.551267 |
3
|
4259
|
|
GO:0003674
|
molecular_function
|
0.567526 |
0.567526 |
4
|
11064
|
|
GO:0007560
|
imaginal disc morphogenesis
|
0.575758 |
0.575758 |
1
|
323
|
|
GO:0043039
|
tRNA aminoacylation
|
0.576471 |
0.576471 |
1
|
49
|
|
GO:0071844
|
cellular component assembly at cellular level
|
0.607267 |
0.607267 |
1
|
568
|
|
GO:0009887
|
organ morphogenesis
|
0.608733 |
0.608733 |
1
|
684
|
|
GO:0007444
|
imaginal disc development
|
0.610821 |
0.610821 |
1
|
467
|
|
GO:0003702
|
RNA polymerase II transcription factor activity
|
0.618056 |
0.618056 |
1
|
267
|
|
GO:0005911
|
cell-cell junction
|
0.623762 |
0.623762 |
1
|
63
|
|
GO:0035151
|
regulation of tube size, open tracheal system
|
0.628571 |
0.628571 |
1
|
44
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.631500 |
0.631500 |
3
|
1535
|
|
GO:0035120
|
post-embryonic appendage morphogenesis
|
0.638095 |
0.638095 |
1
|
268
|
|
GO:0044425
|
membrane part
|
0.640589 |
0.640589 |
1
|
1398
|
|
GO:0006897
|
endocytosis
|
0.655814 |
0.655814 |
1
|
282
|
|
GO:0048563
|
post-embryonic organ morphogenesis
|
0.658283 |
0.658283 |
1
|
323
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.662696 |
0.662696 |
2
|
701
|
|
GO:0044424
|
intracellular part
|
0.665216 |
0.665216 |
3
|
4384
|
|
GO:0007472
|
wing disc morphogenesis
|
0.665796 |
0.665796 |
1
|
255
|
|
GO:0007519
|
skeletal muscle tissue development
|
0.669565 |
0.669565 |
1
|
77
|
|
GO:0035220
|
wing disc development
|
0.680942 |
0.680942 |
1
|
318
|
|
GO:0007528
|
neuromuscular junction development
|
0.684783 |
0.684783 |
1
|
63
|
|
GO:0050808
|
synapse organization
|
0.686567 |
0.686567 |
1
|
92
|
|
GO:0043296
|
apical junction complex
|
0.686567 |
0.686567 |
1
|
46
|
|
GO:0019991
|
septate junction assembly
|
0.694444 |
0.694444 |
1
|
25
|
|
GO:0006399
|
tRNA metabolic process
|
0.705882 |
0.705882 |
1
|
84
|
|
GO:0048747
|
muscle fiber development
|
0.717172 |
0.717172 |
1
|
71
|
|
GO:0016787
|
hydrolase activity
|
0.729944 |
0.729944 |
1
|
1972
|
|
GO:0048856
|
anatomical structure development
|
0.737157 |
0.737157 |
2
|
2265
|
|
GO:0065008
|
regulation of biological quality
|
0.740509 |
0.740509 |
1
|
729
|
|
GO:0032553
|
ribonucleotide binding
|
0.746180 |
0.746180 |
1
|
879
|
|
GO:0048741
|
skeletal muscle fiber development
|
0.750000 |
0.750000 |
1
|
63
|
|
GO:0007399
|
nervous system development
|
0.750147 |
0.750147 |
1
|
848
|
|
GO:0032559
|
adenyl ribonucleotide binding
|
0.757705 |
0.757705 |
1
|
713
|
|
GO:0005622
|
intracellular
|
0.761983 |
0.761983 |
3
|
4734
|
|
GO:0007043
|
cell-cell junction assembly
|
0.765957 |
0.765957 |
1
|
36
|
|
GO:0060429
|
epithelium development
|
0.779194 |
0.779194 |
1
|
295
|
|
GO:0055002
|
striated muscle cell development
|
0.798387 |
0.798387 |
1
|
99
|
|
GO:0017076
|
purine nucleotide binding
|
0.803056 |
0.803056 |
1
|
946
|
|
GO:0044444
|
cytoplasmic part
|
0.809942 |
0.809942 |
1
|
1863
|
|
GO:0016020
|
membrane
|
0.813245 |
0.813245 |
1
|
2122
|
|
GO:0030554
|
adenyl nucleotide binding
|
0.819239 |
0.819239 |
1
|
775
|
|
GO:0010324
|
membrane invagination
|
0.819767 |
0.819767 |
1
|
282
|
|
GO:0044267
|
cellular protein metabolic process
|
0.824560 |
0.824560 |
1
|
1505
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.824951 |
0.824951 |
1
|
1534
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.842767 |
0.842767 |
1
|
402
|
|
GO:0019538
|
protein metabolic process
|
0.855761 |
0.855761 |
1
|
2053
|
|
GO:0022403
|
cell cycle phase
|
0.896183 |
0.896183 |
1
|
587
|
|
GO:0007476
|
imaginal disc-derived wing morphogenesis
|
0.903915 |
0.903915 |
1
|
254
|
|
GO:0005737
|
cytoplasm
|
0.904274 |
0.904274 |
1
|
2378
|
|
GO:0043436
|
oxoacid metabolic process
|
0.925000 |
0.925000 |
1
|
259
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.929175 |
0.929175 |
1
|
879
|
|
GO:0051146
|
striated muscle cell differentiation
|
0.939394 |
0.939394 |
1
|
124
|
|
GO:0003002
|
regionalization
|
0.942272 |
0.942272 |
1
|
506
|
|
GO:0030154
|
cell differentiation
|
0.942825 |
0.942825 |
2
|
1239
|
|
GO:0000279
|
M phase
|
0.945486 |
0.945486 |
1
|
555
|
|
GO:0016043
|
cellular component organization
|
0.947563 |
0.947563 |
2
|
2052
|
|
GO:0035315
|
hair cell differentiation
|
0.956522 |
0.956522 |
1
|
44
|
|
GO:0045216
|
cell-cell junction organization
|
0.957447 |
0.957447 |
1
|
45
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0043297
|
apical junction assembly
|
0.972222 |
0.972222 |
1
|
35
|
|
GO:0035114
|
imaginal disc-derived appendage morphogenesis
|
0.975524 |
0.975524 |
1
|
279
|
|
GO:0007623
|
circadian rhythm
|
0.985507 |
0.985507 |
1
|
68
|
|
GO:0048737
|
imaginal disc-derived appendage development
|
0.986014 |
0.986014 |
1
|
282
|
|
GO:0014706
|
striated muscle tissue development
|
0.989011 |
0.989011 |
1
|
90
|
|
GO:0001883
|
purine nucleoside binding
|
0.992347 |
0.992347 |
1
|
778
|
|
GO:0007424
|
open tracheal system development
|
0.995984 |
0.995984 |
1
|
248
|
|
GO:0002165
|
instar larval or pupal development
|
0.997972 |
0.997972 |
1
|
477
|
|
GO:0005524
|
ATP binding
|
0.998597 |
0.998597 |
1
|
712
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
4
|
12747
|
|
GO:0016876
|
ligase activity, forming aminoacyl-tRNA and related compounds
|
1.00000 |
1.00000 |
1
|
53
|
|
GO:0019752
|
carboxylic acid metabolic process
|
1.00000 |
1.00000 |
1
|
259
|
|
GO:0003700
|
sequence-specific DNA binding transcription factor activity
|
1.00000 |
1.00000 |
1
|
395
|
|
GO:0004812
|
aminoacyl-tRNA ligase activity
|
1.00000 |
1.00000 |
1
|
53
|