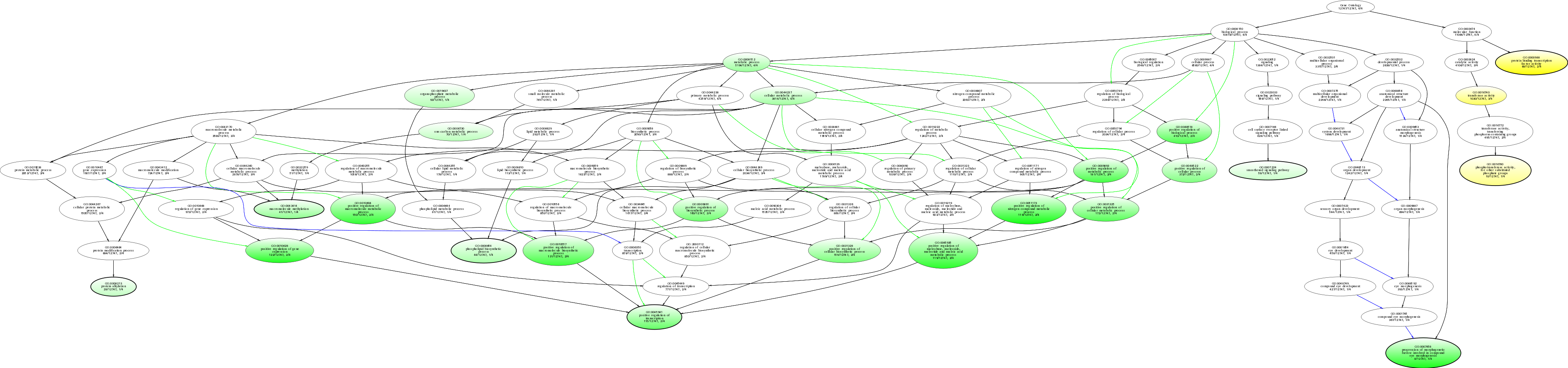
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0000988
|
protein binding transcription factor activity
|
0.000221559 |
0.000221559 |
2
|
68
|
|
GO:0051173
|
positive regulation of nitrogen compound metabolic process
|
0.00308492 |
0.00308492 |
2
|
119
|
|
GO:0007458
|
progression of morphogenetic furrow involved in compound eye morphogenesis
|
0.00353201 |
0.00353201 |
1
|
8
|
|
GO:0010628
|
positive regulation of gene expression
|
0.00401909 |
0.00401909 |
2
|
122
|
|
GO:0045935
|
positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.00419073 |
0.00419073 |
2
|
119
|
|
GO:0010604
|
positive regulation of macromolecule metabolic process
|
0.00501389 |
0.00501389 |
2
|
150
|
|
GO:0010557
|
positive regulation of macromolecule biosynthetic process
|
0.00619133 |
0.00619133 |
2
|
131
|
|
GO:0009893
|
positive regulation of metabolic process
|
0.00664587 |
0.00664587 |
2
|
181
|
|
GO:0048518
|
positive regulation of biological process
|
0.00847745 |
0.00847745 |
2
|
410
|
|
GO:0031325
|
positive regulation of cellular metabolic process
|
0.0108334 |
0.0108334 |
2
|
172
|
|
GO:0016740
|
transferase activity
|
0.0157509 |
0.0157509 |
3
|
1030
|
|
GO:0045941
|
positive regulation of transcription
|
0.0161672 |
0.0161672 |
2
|
115
|
|
GO:0009891
|
positive regulation of biosynthetic process
|
0.0163994 |
0.0163994 |
2
|
160
|
|
GO:0031328
|
positive regulation of cellular biosynthetic process
|
0.0171228 |
0.0171228 |
2
|
159
|
|
GO:0048522
|
positive regulation of cellular process
|
0.0177709 |
0.0177709 |
2
|
372
|
|
GO:0016780
|
phosphotransferase activity, for other substituted phosphate groups
|
0.0400360 |
0.0400360 |
1
|
10
|
|
GO:0043414
|
macromolecule methylation
|
0.0420026 |
0.0420026 |
1
|
41
|
|
GO:0008152
|
metabolic process
|
0.0572674 |
0.0572674 |
4
|
5194
|
|
GO:0044237
|
cellular metabolic process
|
0.0580681 |
0.0580681 |
4
|
3814
|
|
GO:0008654
|
phospholipid biosynthetic process
|
0.0623045 |
0.0623045 |
1
|
44
|
|
GO:0019637
|
organophosphate metabolic process
|
0.0697400 |
0.0697400 |
1
|
93
|
|
GO:0006730
|
one-carbon metabolic process
|
0.0709560 |
0.0709560 |
1
|
70
|
|
GO:0008213
|
protein alkylation
|
0.0802531 |
0.0802531 |
1
|
28
|
|
GO:0007224
|
smoothened signaling pathway
|
0.0929487 |
0.0929487 |
1
|
58
|
|
GO:0005102
|
receptor binding
|
0.100232 |
0.100232 |
1
|
173
|
|
GO:0043412
|
macromolecule modification
|
0.105638 |
0.105638 |
2
|
724
|
|
GO:0001071
|
nucleic acid binding transcription factor activity
|
0.135356 |
0.135356 |
1
|
395
|
|
GO:0016563
|
transcription activator activity
|
0.141204 |
0.141204 |
1
|
61
|
|
GO:0008610
|
lipid biosynthetic process
|
0.142505 |
0.142505 |
1
|
113
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.145592 |
0.145592 |
2
|
905
|
|
GO:0009987
|
cellular process
|
0.145754 |
0.145754 |
4
|
6560
|
|
GO:0030528
|
transcription regulator activity
|
0.147289 |
0.147289 |
1
|
432
|
|
GO:0003824
|
catalytic activity
|
0.147500 |
0.147500 |
3
|
4106
|
|
GO:0006350
|
transcription
|
0.150290 |
0.150290 |
2
|
859
|
|
GO:0004307
|
ethanolaminephosphotransferase activity
|
0.166667 |
0.166667 |
1
|
1
|
|
GO:0044255
|
cellular lipid metabolic process
|
0.170259 |
0.170259 |
1
|
178
|
|
GO:0016570
|
histone modification
|
0.170528 |
0.170528 |
1
|
61
|
|
GO:0009058
|
biosynthetic process
|
0.181873 |
0.181873 |
3
|
2090
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.194907 |
0.194907 |
2
|
1056
|
|
GO:0006464
|
protein modification process
|
0.196094 |
0.196094 |
2
|
684
|
|
GO:0019222
|
regulation of metabolic process
|
0.196929 |
0.196929 |
2
|
1242
|
|
GO:0003677
|
DNA binding
|
0.197185 |
0.197185 |
2
|
824
|
|
GO:0050789
|
regulation of biological process
|
0.198789 |
0.198789 |
2
|
2246
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.203932 |
0.203932 |
2
|
903
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.208706 |
0.208706 |
2
|
850
|
|
GO:0042800
|
histone methyltransferase activity (H3-K4 specific)
|
0.222222 |
0.222222 |
1
|
4
|
|
GO:0010468
|
regulation of gene expression
|
0.239905 |
0.239905 |
2
|
973
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.240283 |
0.240283 |
2
|
1039
|
|
GO:0065007
|
biological regulation
|
0.244964 |
0.244964 |
2
|
2548
|
|
GO:0006629
|
lipid metabolic process
|
0.247384 |
0.247384 |
1
|
292
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.252773 |
0.252773 |
2
|
1110
|
|
GO:0003676
|
nucleic acid binding
|
0.253252 |
0.253252 |
2
|
1855
|
|
GO:0048232
|
male gamete generation
|
0.254994 |
0.254994 |
1
|
217
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.263629 |
0.263629 |
2
|
850
|
|
GO:0005700
|
polytene chromosome
|
0.276650 |
0.276650 |
1
|
109
|
|
GO:0016741
|
transferase activity, transferring one-carbon groups
|
0.289962 |
0.289962 |
1
|
111
|
|
GO:0008276
|
protein methyltransferase activity
|
0.304762 |
0.304762 |
1
|
32
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.304837 |
0.304837 |
2
|
888
|
|
GO:0016571
|
histone methylation
|
0.308824 |
0.308824 |
1
|
21
|
|
GO:0016278
|
lysine N-methyltransferase activity
|
0.310345 |
0.310345 |
1
|
18
|
|
GO:0008170
|
N-methyltransferase activity
|
0.314286 |
0.314286 |
1
|
33
|
|
GO:0005694
|
chromosome
|
0.321108 |
0.321108 |
1
|
394
|
|
GO:0005488
|
binding
|
0.327389 |
0.327389 |
3
|
5641
|
|
GO:0000003
|
reproduction
|
0.332995 |
0.332995 |
1
|
1022
|
|
GO:0022414
|
reproductive process
|
0.332995 |
0.332995 |
1
|
1022
|
|
GO:0016568
|
chromatin modification
|
0.344322 |
0.344322 |
1
|
94
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.346544 |
0.346544 |
2
|
888
|
|
GO:0005623
|
cell
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0044464
|
cell part
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0043226
|
organelle
|
0.353651 |
0.353651 |
2
|
3537
|
|
GO:0044249
|
cellular biosynthetic process
|
0.353662 |
0.353662 |
3
|
2034
|
|
GO:0051276
|
chromosome organization
|
0.358714 |
0.358714 |
1
|
424
|
|
GO:0050794
|
regulation of cellular process
|
0.359544 |
0.359544 |
2
|
2034
|
|
GO:0001882
|
nucleoside binding
|
0.361738 |
0.361738 |
1
|
784
|
|
GO:0032501
|
multicellular organismal process
|
0.369984 |
0.369984 |
2
|
3315
|
|
GO:0009887
|
organ morphogenesis
|
0.374384 |
0.374384 |
1
|
684
|
|
GO:0005634
|
nucleus
|
0.408695 |
0.408695 |
2
|
1826
|
|
GO:0044267
|
cellular protein metabolic process
|
0.410493 |
0.410493 |
2
|
1505
|
|
GO:0023052
|
signaling
|
0.423150 |
0.423150 |
1
|
1364
|
|
GO:0006644
|
phospholipid metabolic process
|
0.428571 |
0.428571 |
1
|
81
|
|
GO:0007423
|
sensory organ development
|
0.438003 |
0.438003 |
1
|
544
|
|
GO:0044238
|
primary metabolic process
|
0.451973 |
0.451973 |
4
|
4259
|
|
GO:0006793
|
phosphorus metabolic process
|
0.467690 |
0.467690 |
1
|
556
|
|
GO:0044281
|
small molecule metabolic process
|
0.469486 |
0.469486 |
1
|
761
|
|
GO:0016772
|
transferase activity, transferring phosphorus-containing groups
|
0.470860 |
0.470860 |
2
|
495
|
|
GO:0045449
|
regulation of transcription
|
0.472005 |
0.472005 |
2
|
771
|
|
GO:0048609
|
multicellular organismal reproductive process
|
0.480389 |
0.480389 |
1
|
937
|
|
GO:0032504
|
multicellular organism reproduction
|
0.480389 |
0.480389 |
1
|
937
|
|
GO:0008150
|
biological_process
|
0.481028 |
0.481028 |
4
|
10616
|
|
GO:0000166
|
nucleotide binding
|
0.504833 |
0.504833 |
1
|
1178
|
|
GO:0043565
|
sequence-specific DNA binding
|
0.511614 |
0.511614 |
1
|
248
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.529952 |
0.529952 |
2
|
2093
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.534584 |
0.534584 |
2
|
1623
|
|
GO:0048592
|
eye morphogenesis
|
0.542069 |
0.542069 |
1
|
393
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.543703 |
0.543703 |
3
|
2878
|
|
GO:0010467
|
gene expression
|
0.547191 |
0.547191 |
2
|
1907
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.551885 |
0.551885 |
2
|
1789
|
|
GO:0008757
|
S-adenosylmethionine-dependent methyltransferase activity
|
0.552381 |
0.552381 |
1
|
58
|
|
GO:0006468
|
protein phosphorylation
|
0.559053 |
0.559053 |
1
|
277
|
|
GO:0016279
|
protein-lysine N-methyltransferase activity
|
0.562500 |
0.562500 |
1
|
18
|
|
GO:0003714
|
transcription corepressor activity
|
0.565408 |
0.565408 |
1
|
23
|
|
GO:0003674
|
molecular_function
|
0.567526 |
0.567526 |
4
|
11064
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.573530 |
0.573530 |
1
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.574428 |
0.574428 |
1
|
1227
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.587518 |
0.587518 |
1
|
2108
|
|
GO:0017169
|
CDP-alcohol phosphatidyltransferase activity
|
0.600000 |
0.600000 |
1
|
6
|
|
GO:0043167
|
ion binding
|
0.603911 |
0.603911 |
1
|
1498
|
|
GO:0043170
|
macromolecule metabolic process
|
0.635446 |
0.635446 |
3
|
3588
|
|
GO:0005575
|
cellular_component
|
0.637024 |
0.637024 |
3
|
8817
|
|
GO:0032774
|
RNA biosynthetic process
|
0.642295 |
0.642295 |
1
|
703
|
|
GO:0006325
|
chromatin organization
|
0.643868 |
0.643868 |
1
|
273
|
|
GO:0043229
|
intracellular organelle
|
0.646541 |
0.646541 |
2
|
3530
|
|
GO:0016569
|
covalent chromatin modification
|
0.648936 |
0.648936 |
1
|
61
|
|
GO:0019538
|
protein metabolic process
|
0.649882 |
0.649882 |
2
|
2053
|
|
GO:0007166
|
cell surface receptor linked signaling pathway
|
0.652720 |
0.652720 |
1
|
624
|
|
GO:0043227
|
membrane-bounded organelle
|
0.653325 |
0.653325 |
2
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.653801 |
0.653801 |
2
|
2856
|
|
GO:0042054
|
histone methyltransferase activity
|
0.656250 |
0.656250 |
1
|
21
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.661861 |
0.661861 |
1
|
1562
|
|
GO:0005515
|
protein binding
|
0.665786 |
0.665786 |
1
|
1726
|
|
GO:0032502
|
developmental process
|
0.681102 |
0.681102 |
1
|
2638
|
|
GO:0006479
|
protein methylation
|
0.682927 |
0.682927 |
1
|
28
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.682990 |
0.682990 |
2
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.685110 |
0.685110 |
2
|
1959
|
|
GO:0004674
|
protein serine/threonine kinase activity
|
0.686207 |
0.686207 |
1
|
199
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.686903 |
0.686903 |
1
|
686
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0016020
|
membrane
|
0.715875 |
0.715875 |
1
|
2122
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.724105 |
0.724105 |
1
|
1517
|
|
GO:0032259
|
methylation
|
0.728571 |
0.728571 |
1
|
51
|
|
GO:0004672
|
protein kinase activity
|
0.728643 |
0.728643 |
1
|
290
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.736132 |
0.736132 |
2
|
1535
|
|
GO:0032553
|
ribonucleotide binding
|
0.746180 |
0.746180 |
1
|
879
|
|
GO:0003713
|
transcription coactivator activity
|
0.753731 |
0.753731 |
1
|
34
|
|
GO:0032559
|
adenyl ribonucleotide binding
|
0.757705 |
0.757705 |
1
|
713
|
|
GO:0016310
|
phosphorylation
|
0.758993 |
0.758993 |
1
|
422
|
|
GO:0006996
|
organelle organization
|
0.779169 |
0.779169 |
1
|
1182
|
|
GO:0008270
|
zinc ion binding
|
0.781440 |
0.781440 |
1
|
901
|
|
GO:0044424
|
intracellular part
|
0.793631 |
0.793631 |
2
|
4384
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0016070
|
RNA metabolic process
|
0.798742 |
0.798742 |
1
|
1191
|
|
GO:0017076
|
purine nucleotide binding
|
0.803056 |
0.803056 |
1
|
946
|
|
GO:0001745
|
compound eye morphogenesis
|
0.811947 |
0.811947 |
1
|
367
|
|
GO:0030554
|
adenyl nucleotide binding
|
0.819239 |
0.819239 |
1
|
775
|
|
GO:0001654
|
eye development
|
0.841912 |
0.841912 |
1
|
458
|
|
GO:0018024
|
histone-lysine N-methyltransferase activity
|
0.857143 |
0.857143 |
1
|
18
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0019953
|
sexual reproduction
|
0.859100 |
0.859100 |
1
|
878
|
|
GO:0005622
|
intracellular
|
0.859425 |
0.859425 |
2
|
4734
|
|
GO:0007276
|
gamete generation
|
0.883697 |
0.883697 |
1
|
851
|
|
GO:0007275
|
multicellular organismal development
|
0.887792 |
0.887792 |
1
|
2296
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.900589 |
0.900589 |
1
|
620
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.929175 |
0.929175 |
1
|
879
|
|
GO:0016773
|
phosphotransferase activity, alcohol group as acceptor
|
0.930356 |
0.930356 |
1
|
364
|
|
GO:0048749
|
compound eye development
|
0.932314 |
0.932314 |
1
|
427
|
|
GO:0008168
|
methyltransferase activity
|
0.945946 |
0.945946 |
1
|
105
|
|
GO:0016301
|
kinase activity
|
0.958696 |
0.958696 |
1
|
394
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.965643 |
0.965643 |
1
|
701
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0001883
|
purine nucleoside binding
|
0.992347 |
0.992347 |
1
|
778
|
|
GO:0007283
|
spermatogenesis
|
0.995392 |
0.995392 |
1
|
216
|
|
GO:0005524
|
ATP binding
|
0.998597 |
0.998597 |
1
|
712
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
4
|
12747
|
|
GO:0000989
|
transcription factor binding transcription factor activity
|
1.00000 |
1.00000 |
2
|
68
|
|
GO:0003712
|
transcription cofactor activity
|
1.00000 |
1.00000 |
2
|
68
|
|
GO:0003700
|
sequence-specific DNA binding transcription factor activity
|
1.00000 |
1.00000 |
1
|
395
|
|
GO:0006796
|
phosphate metabolic process
|
1.00000 |
1.00000 |
1
|
556
|