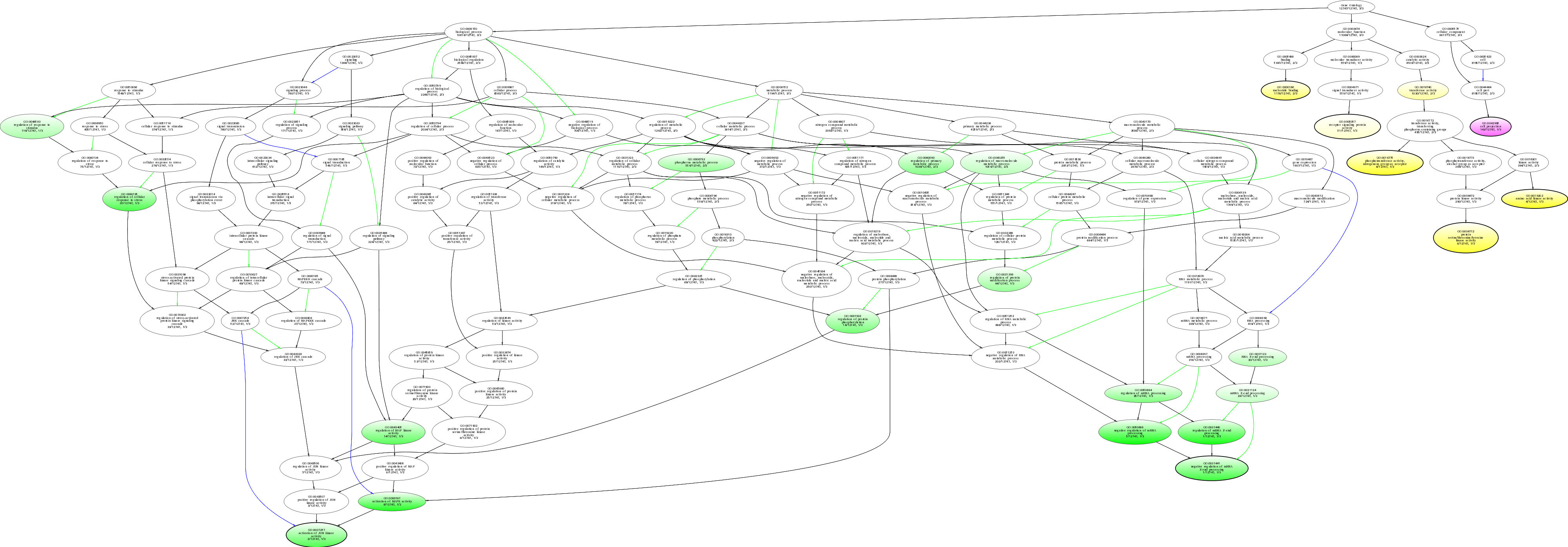
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0050686
|
negative regulation of mRNA processing
|
0.00986193 |
0.00986193 |
1
|
5
|
|
GO:0031440
|
regulation of mRNA 3'-end processing
|
0.0111111 |
0.0111111 |
1
|
1
|
|
GO:0000187
|
activation of MAPK activity
|
0.0185185 |
0.0185185 |
1
|
6
|
|
GO:0019202
|
amino acid kinase activity
|
0.0202271 |
0.0202271 |
1
|
4
|
|
GO:0016775
|
phosphotransferase activity, nitrogenous group as acceptor
|
0.0241197 |
0.0241197 |
1
|
6
|
|
GO:0004712
|
protein serine/threonine/tyrosine kinase activity
|
0.0275862 |
0.0275862 |
1
|
8
|
|
GO:0031441
|
negative regulation of mRNA 3'-end processing
|
0.0312500 |
0.0312500 |
1
|
1
|
|
GO:0080135
|
regulation of cellular response to stress
|
0.0319322 |
0.0319322 |
1
|
35
|
|
GO:0007257
|
activation of JUN kinase activity
|
0.0357143 |
0.0357143 |
1
|
2
|
|
GO:0043405
|
regulation of MAP kinase activity
|
0.0408163 |
0.0408163 |
1
|
14
|
|
GO:0000166
|
nucleotide binding
|
0.0435799 |
0.0435799 |
2
|
1178
|
|
GO:0042995
|
cell projection
|
0.0456371 |
0.0456371 |
1
|
143
|
|
GO:0001932
|
regulation of protein phosphorylation
|
0.0459770 |
0.0459770 |
1
|
16
|
|
GO:0050684
|
regulation of mRNA processing
|
0.0542571 |
0.0542571 |
1
|
65
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.0562295 |
0.0562295 |
2
|
1039
|
|
GO:0006793
|
phosphorus metabolic process
|
0.0574888 |
0.0574888 |
2
|
556
|
|
GO:0031399
|
regulation of protein modification process
|
0.0579710 |
0.0579710 |
1
|
44
|
|
GO:0016740
|
transferase activity
|
0.0628812 |
0.0628812 |
2
|
1030
|
|
GO:0048583
|
regulation of response to stimulus
|
0.0699916 |
0.0699916 |
1
|
114
|
|
GO:0031123
|
RNA 3'-end processing
|
0.0724638 |
0.0724638 |
1
|
30
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.0800539 |
0.0800539 |
2
|
1056
|
|
GO:0031124
|
mRNA 3'-end processing
|
0.0875000 |
0.0875000 |
1
|
28
|
|
GO:0005057
|
receptor signaling protein activity
|
0.0912343 |
0.0912343 |
1
|
51
|
|
GO:0051246
|
regulation of protein metabolic process
|
0.102375 |
0.102375 |
1
|
155
|
|
GO:0032268
|
regulation of cellular protein metabolic process
|
0.102432 |
0.102432 |
1
|
128
|
|
GO:0019222
|
regulation of metabolic process
|
0.114129 |
0.114129 |
2
|
1242
|
|
GO:0051172
|
negative regulation of nitrogen compound metabolic process
|
0.114943 |
0.114943 |
1
|
250
|
|
GO:0050789
|
regulation of biological process
|
0.115315 |
0.115315 |
2
|
2246
|
|
GO:0008152
|
metabolic process
|
0.117083 |
0.117083 |
3
|
5194
|
|
GO:0080134
|
regulation of response to stress
|
0.117925 |
0.117925 |
1
|
75
|
|
GO:0044237
|
cellular metabolic process
|
0.118315 |
0.118315 |
3
|
3814
|
|
GO:0023014
|
signal transmission via phosphorylation event
|
0.126316 |
0.126316 |
1
|
96
|
|
GO:0065009
|
regulation of molecular function
|
0.126812 |
0.126812 |
1
|
167
|
|
GO:0010627
|
regulation of intracellular protein kinase cascade
|
0.132075 |
0.132075 |
1
|
49
|
|
GO:0045934
|
negative regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.135355 |
0.135355 |
1
|
250
|
|
GO:0051174
|
regulation of phosphorus metabolic process
|
0.138139 |
0.138139 |
1
|
76
|
|
GO:0023051
|
regulation of signaling process
|
0.139523 |
0.139523 |
1
|
171
|
|
GO:0043506
|
regulation of JUN kinase activity
|
0.142857 |
0.142857 |
1
|
7
|
|
GO:0060089
|
molecular transducer activity
|
0.144056 |
0.144056 |
1
|
559
|
|
GO:0065007
|
biological regulation
|
0.145142 |
0.145142 |
2
|
2548
|
|
GO:0051716
|
cellular response to stimulus
|
0.150190 |
0.150190 |
1
|
374
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.150277 |
0.150277 |
2
|
1110
|
|
GO:0032502
|
developmental process
|
0.154531 |
0.154531 |
2
|
2638
|
|
GO:0051253
|
negative regulation of RNA metabolic process
|
0.155744 |
0.155744 |
1
|
202
|
|
GO:0016604
|
nuclear body
|
0.157692 |
0.157692 |
1
|
41
|
|
GO:0004702
|
receptor signaling protein serine/threonine kinase activity
|
0.169014 |
0.169014 |
1
|
36
|
|
GO:0031098
|
stress-activated protein kinase signaling cascade
|
0.170886 |
0.170886 |
1
|
54
|
|
GO:0048519
|
negative regulation of biological process
|
0.186553 |
0.186553 |
1
|
706
|
|
GO:0010605
|
negative regulation of macromolecule metabolic process
|
0.187925 |
0.187925 |
1
|
356
|
|
GO:0009892
|
negative regulation of metabolic process
|
0.193590 |
0.193590 |
1
|
372
|
|
GO:0023046
|
signaling process
|
0.199779 |
0.199779 |
1
|
760
|
|
GO:0031974
|
membrane-enclosed lumen
|
0.204405 |
0.204405 |
1
|
647
|
|
GO:0006396
|
RNA processing
|
0.214286 |
0.214286 |
1
|
414
|
|
GO:0071902
|
positive regulation of protein serine/threonine kinase activity
|
0.216216 |
0.216216 |
1
|
8
|
|
GO:0031324
|
negative regulation of cellular metabolic process
|
0.219618 |
0.219618 |
1
|
319
|
|
GO:0050794
|
regulation of cellular process
|
0.224039 |
0.224039 |
2
|
2034
|
|
GO:0050790
|
regulation of catalytic activity
|
0.224781 |
0.224781 |
1
|
149
|
|
GO:0016772
|
transferase activity, transferring phosphorus-containing groups
|
0.230717 |
0.230717 |
2
|
495
|
|
GO:0032501
|
multicellular organismal process
|
0.231607 |
0.231607 |
2
|
3315
|
|
GO:0009987
|
cellular process
|
0.235914 |
0.235914 |
3
|
6560
|
|
GO:0035466
|
regulation of signaling pathway
|
0.237323 |
0.237323 |
1
|
324
|
|
GO:0007399
|
nervous system development
|
0.249853 |
0.249853 |
2
|
848
|
|
GO:0043507
|
positive regulation of JUN kinase activity
|
0.250000 |
0.250000 |
1
|
3
|
|
GO:0007417
|
central nervous system development
|
0.252905 |
0.252905 |
1
|
230
|
|
GO:0019220
|
regulation of phosphate metabolic process
|
0.254910 |
0.254910 |
1
|
76
|
|
GO:0001882
|
nucleoside binding
|
0.258670 |
0.258670 |
1
|
784
|
|
GO:0048523
|
negative regulation of cellular process
|
0.262869 |
0.262869 |
1
|
635
|
|
GO:0044428
|
nuclear part
|
0.271419 |
0.271419 |
1
|
830
|
|
GO:0051347
|
positive regulation of transferase activity
|
0.271739 |
0.271739 |
1
|
25
|
|
GO:0016071
|
mRNA metabolic process
|
0.284635 |
0.284635 |
1
|
339
|
|
GO:0043233
|
organelle lumen
|
0.291360 |
0.291360 |
1
|
634
|
|
GO:0070013
|
intracellular organelle lumen
|
0.293247 |
0.293247 |
1
|
634
|
|
GO:0042325
|
regulation of phosphorylation
|
0.296124 |
0.296124 |
1
|
69
|
|
GO:0003729
|
mRNA binding
|
0.302147 |
0.302147 |
1
|
197
|
|
GO:0007243
|
intracellular protein kinase cascade
|
0.304762 |
0.304762 |
1
|
96
|
|
GO:0003824
|
catalytic activity
|
0.310936 |
0.310936 |
2
|
4106
|
|
GO:0009966
|
regulation of signal transduction
|
0.312044 |
0.312044 |
1
|
171
|
|
GO:0044451
|
nucleoplasm part
|
0.313253 |
0.313253 |
1
|
260
|
|
GO:0005575
|
cellular_component
|
0.330897 |
0.330897 |
3
|
8817
|
|
GO:0023052
|
signaling
|
0.338079 |
0.338079 |
1
|
1364
|
|
GO:0005654
|
nucleoplasm
|
0.340964 |
0.340964 |
1
|
283
|
|
GO:0003723
|
RNA binding
|
0.351482 |
0.351482 |
1
|
652
|
|
GO:0051338
|
regulation of transferase activity
|
0.355705 |
0.355705 |
1
|
53
|
|
GO:0043412
|
macromolecule modification
|
0.362896 |
0.362896 |
1
|
724
|
|
GO:0050896
|
response to stimulus
|
0.376795 |
0.376795 |
1
|
1548
|
|
GO:0071900
|
regulation of protein serine/threonine kinase activity
|
0.377358 |
0.377358 |
1
|
20
|
|
GO:0006950
|
response to stress
|
0.390827 |
0.390827 |
1
|
605
|
|
GO:0033554
|
cellular response to stress
|
0.397122 |
0.397122 |
1
|
276
|
|
GO:0043085
|
positive regulation of catalytic activity
|
0.405063 |
0.405063 |
1
|
64
|
|
GO:0004054
|
arginine kinase activity
|
0.428571 |
0.428571 |
1
|
3
|
|
GO:0023034
|
intracellular signaling pathway
|
0.430962 |
0.430962 |
1
|
412
|
|
GO:0048731
|
system development
|
0.432376 |
0.432376 |
2
|
1696
|
|
GO:0048666
|
neuron development
|
0.433702 |
0.433702 |
1
|
471
|
|
GO:0043408
|
regulation of MAPKKK cascade
|
0.435294 |
0.435294 |
1
|
37
|
|
GO:0044093
|
positive regulation of molecular function
|
0.437126 |
0.437126 |
1
|
73
|
|
GO:0007275
|
multicellular organismal development
|
0.442066 |
0.442066 |
2
|
2296
|
|
GO:0048869
|
cellular developmental process
|
0.442161 |
0.442161 |
1
|
1276
|
|
GO:0030182
|
neuron differentiation
|
0.442292 |
0.442292 |
1
|
548
|
|
GO:0006464
|
protein modification process
|
0.443005 |
0.443005 |
1
|
684
|
|
GO:0007165
|
signal transduction
|
0.443656 |
0.443656 |
1
|
548
|
|
GO:0070302
|
regulation of stress-activated protein kinase signaling cascade
|
0.458333 |
0.458333 |
1
|
33
|
|
GO:0033674
|
positive regulation of kinase activity
|
0.471698 |
0.471698 |
1
|
25
|
|
GO:0045860
|
positive regulation of protein kinase activity
|
0.471698 |
0.471698 |
1
|
25
|
|
GO:0031981
|
nuclear lumen
|
0.475277 |
0.475277 |
1
|
471
|
|
GO:0044446
|
intracellular organelle part
|
0.493044 |
0.493044 |
1
|
2162
|
|
GO:0035556
|
intracellular signal transduction
|
0.499208 |
0.499208 |
1
|
315
|
|
GO:0004708
|
MAP kinase kinase activity
|
0.500000 |
0.500000 |
1
|
4
|
|
GO:0008545
|
JUN kinase kinase activity
|
0.500000 |
0.500000 |
1
|
2
|
|
GO:0043005
|
neuron projection
|
0.503497 |
0.503497 |
1
|
72
|
|
GO:0005488
|
binding
|
0.514777 |
0.514777 |
2
|
5641
|
|
GO:0015030
|
Cajal body
|
0.536585 |
0.536585 |
1
|
22
|
|
GO:0003676
|
nucleic acid binding
|
0.549587 |
0.549587 |
1
|
1855
|
|
GO:0006468
|
protein phosphorylation
|
0.559053 |
0.559053 |
1
|
277
|
|
GO:0043406
|
positive regulation of MAP kinase activity
|
0.571429 |
0.571429 |
1
|
8
|
|
GO:0044422
|
organelle part
|
0.572743 |
0.572743 |
1
|
2176
|
|
GO:0016310
|
phosphorylation
|
0.575740 |
0.575740 |
2
|
422
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0046328
|
regulation of JNK cascade
|
0.589286 |
0.589286 |
1
|
33
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.617799 |
0.617799 |
1
|
905
|
|
GO:0016301
|
kinase activity
|
0.633223 |
0.633223 |
2
|
394
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0016070
|
RNA metabolic process
|
0.656488 |
0.656488 |
1
|
1191
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.658716 |
0.658716 |
1
|
1789
|
|
GO:0004674
|
protein serine/threonine kinase activity
|
0.686207 |
0.686207 |
1
|
199
|
|
GO:0044267
|
cellular protein metabolic process
|
0.686538 |
0.686538 |
1
|
1505
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.686903 |
0.686903 |
1
|
686
|
|
GO:0030424
|
axon
|
0.694444 |
0.694444 |
1
|
50
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.699519 |
0.699519 |
1
|
903
|
|
GO:0022008
|
neurogenesis
|
0.700310 |
0.700310 |
1
|
632
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0007254
|
JNK cascade
|
0.702703 |
0.702703 |
1
|
52
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.703351 |
0.703351 |
2
|
2878
|
|
GO:0048468
|
cell development
|
0.705715 |
0.705715 |
1
|
1042
|
|
GO:0019538
|
protein metabolic process
|
0.724907 |
0.724907 |
1
|
2053
|
|
GO:0006397
|
mRNA processing
|
0.734411 |
0.734411 |
1
|
318
|
|
GO:0048856
|
anatomical structure development
|
0.737157 |
0.737157 |
2
|
2265
|
|
GO:0010468
|
regulation of gene expression
|
0.739954 |
0.739954 |
1
|
973
|
|
GO:0032559
|
adenyl ribonucleotide binding
|
0.757705 |
0.757705 |
1
|
713
|
|
GO:0000165
|
MAPKKK cascade
|
0.760417 |
0.760417 |
1
|
73
|
|
GO:0043549
|
regulation of kinase activity
|
0.768116 |
0.768116 |
1
|
53
|
|
GO:0043170
|
macromolecule metabolic process
|
0.772351 |
0.772351 |
2
|
3588
|
|
GO:0010467
|
gene expression
|
0.780571 |
0.780571 |
1
|
1907
|
|
GO:0043226
|
organelle
|
0.785296 |
0.785296 |
1
|
3537
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.787269 |
0.787269 |
1
|
2093
|
|
GO:0005623
|
cell
|
0.787322 |
0.787322 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.787322 |
0.787322 |
2
|
6195
|
|
GO:0043229
|
intracellular organelle
|
0.804100 |
0.804100 |
1
|
3530
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0030554
|
adenyl nucleotide binding
|
0.819239 |
0.819239 |
1
|
775
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.873851 |
0.873851 |
1
|
1959
|
|
GO:0044238
|
primary metabolic process
|
0.914505 |
0.914505 |
2
|
4259
|
|
GO:0044424
|
intracellular part
|
0.914575 |
0.914575 |
1
|
4384
|
|
GO:0004672
|
protein kinase activity
|
0.926864 |
0.926864 |
1
|
290
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.929175 |
0.929175 |
1
|
879
|
|
GO:0016773
|
phosphotransferase activity, alcohol group as acceptor
|
0.930356 |
0.930356 |
1
|
364
|
|
GO:0032553
|
ribonucleotide binding
|
0.935736 |
0.935736 |
1
|
879
|
|
GO:0005622
|
intracellular
|
0.944411 |
0.944411 |
1
|
4734
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0017076
|
purine nucleotide binding
|
0.961347 |
0.961347 |
1
|
946
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0001883
|
purine nucleoside binding
|
0.992347 |
0.992347 |
1
|
778
|
|
GO:0005524
|
ATP binding
|
0.998597 |
0.998597 |
1
|
712
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0004871
|
signal transducer activity
|
1.00000 |
1.00000 |
1
|
559
|
|
GO:0006796
|
phosphate metabolic process
|
1.00000 |
1.00000 |
2
|
556
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|
|
GO:0045859
|
regulation of protein kinase activity
|
1.00000 |
1.00000 |
1
|
53
|