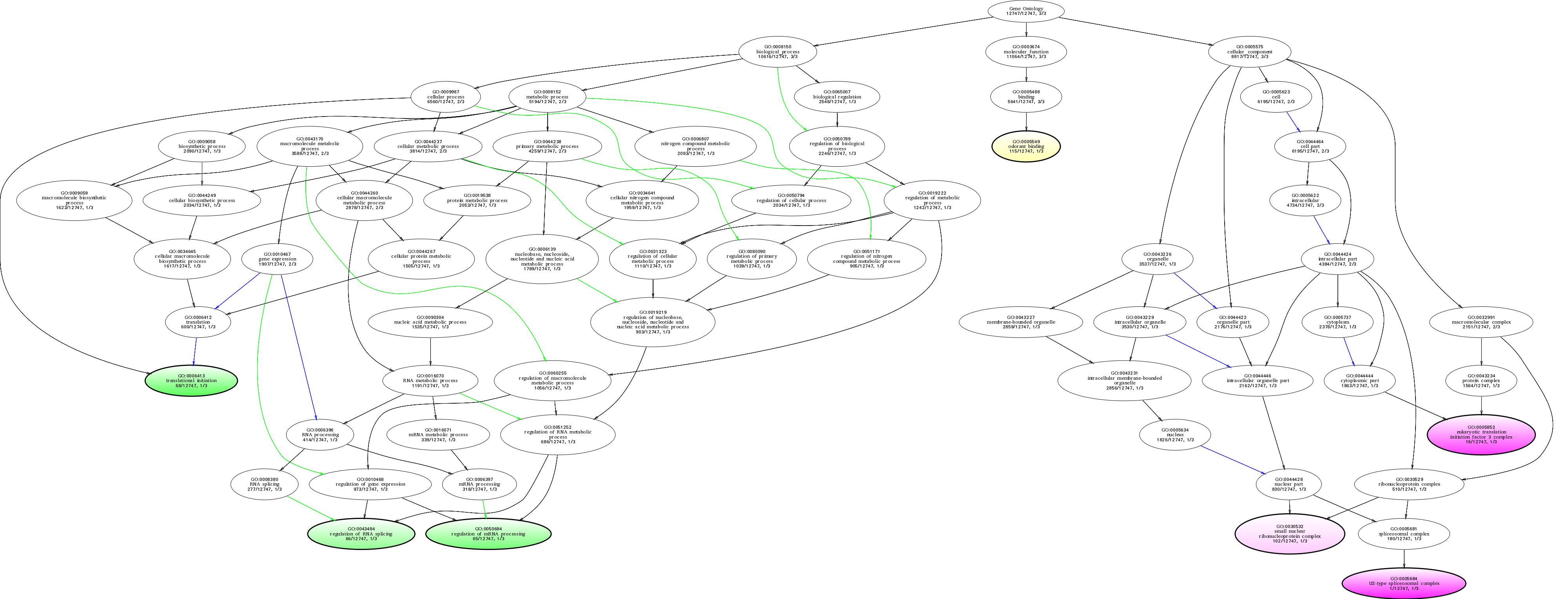
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0005684
|
U2-type spliceosomal complex
|
0.00555556 |
0.00555556 |
1
|
1
|
|
GO:0005852
|
eukaryotic translation initiation factor 3 complex
|
0.00565971 |
0.00565971 |
1
|
16
|
|
GO:0006413
|
translational initiation
|
0.0206258 |
0.0206258 |
1
|
68
|
|
GO:0050684
|
regulation of mRNA processing
|
0.0542571 |
0.0542571 |
1
|
65
|
|
GO:0043484
|
regulation of RNA splicing
|
0.0567498 |
0.0567498 |
1
|
66
|
|
GO:0005549
|
odorant binding
|
0.0599314 |
0.0599314 |
1
|
115
|
|
GO:0030532
|
small nuclear ribonucleoprotein complex
|
0.0949721 |
0.0949721 |
1
|
102
|
|
GO:0008135
|
translation factor activity, nucleic acid binding
|
0.127301 |
0.127301 |
1
|
83
|
|
GO:0005488
|
binding
|
0.132501 |
0.132501 |
3
|
5641
|
|
GO:0032991
|
macromolecular complex
|
0.149479 |
0.149479 |
2
|
2151
|
|
GO:0005682
|
U5 snRNP
|
0.156863 |
0.156863 |
1
|
16
|
|
GO:0005681
|
spliceosomal complex
|
0.167598 |
0.167598 |
1
|
180
|
|
GO:0003008
|
system process
|
0.200302 |
0.200302 |
1
|
664
|
|
GO:0030529
|
ribonucleoprotein complex
|
0.210589 |
0.210589 |
1
|
510
|
|
GO:0000380
|
alternative nuclear mRNA splicing, via spliceosome
|
0.218045 |
0.218045 |
1
|
58
|
|
GO:0005829
|
cytosol
|
0.228127 |
0.228127 |
1
|
425
|
|
GO:0048024
|
regulation of nuclear mRNA splicing, via spliceosome
|
0.237918 |
0.237918 |
1
|
64
|
|
GO:0044237
|
cellular metabolic process
|
0.241038 |
0.241038 |
2
|
3814
|
|
GO:0044428
|
nuclear part
|
0.271419 |
0.271419 |
1
|
830
|
|
GO:0010467
|
gene expression
|
0.282416 |
0.282416 |
2
|
1907
|
|
GO:0016071
|
mRNA metabolic process
|
0.284635 |
0.284635 |
1
|
339
|
|
GO:0003729
|
mRNA binding
|
0.302147 |
0.302147 |
1
|
197
|
|
GO:0005575
|
cellular_component
|
0.330897 |
0.330897 |
3
|
8817
|
|
GO:0003723
|
RNA binding
|
0.351482 |
0.351482 |
1
|
652
|
|
GO:0019222
|
regulation of metabolic process
|
0.376562 |
0.376562 |
1
|
1242
|
|
GO:0051234
|
establishment of localization
|
0.377001 |
0.377001 |
1
|
1549
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.381695 |
0.381695 |
1
|
905
|
|
GO:0006412
|
translation
|
0.382593 |
0.382593 |
1
|
600
|
|
GO:0006396
|
RNA processing
|
0.382740 |
0.382740 |
1
|
414
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.408435 |
0.408435 |
2
|
2878
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.418200 |
0.418200 |
1
|
1039
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.429576 |
0.429576 |
1
|
1110
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.440308 |
0.440308 |
1
|
686
|
|
GO:0051179
|
localization
|
0.445834 |
0.445834 |
1
|
1896
|
|
GO:0007600
|
sensory perception
|
0.448657 |
0.448657 |
1
|
284
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.451726 |
0.451726 |
1
|
903
|
|
GO:0043170
|
macromolecule metabolic process
|
0.477159 |
0.477159 |
2
|
3588
|
|
GO:0008152
|
metabolic process
|
0.483893 |
0.483893 |
2
|
5194
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.486014 |
0.486014 |
1
|
1056
|
|
GO:0044424
|
intracellular part
|
0.500760 |
0.500760 |
2
|
4384
|
|
GO:0000166
|
nucleotide binding
|
0.504833 |
0.504833 |
1
|
1178
|
|
GO:0050789
|
regulation of biological process
|
0.509927 |
0.509927 |
1
|
2246
|
|
GO:0050794
|
regulation of cellular process
|
0.518911 |
0.518911 |
1
|
2034
|
|
GO:0065007
|
biological regulation
|
0.561089 |
0.561089 |
1
|
2548
|
|
GO:0044422
|
organelle part
|
0.572743 |
0.572743 |
1
|
2176
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0005622
|
intracellular
|
0.583919 |
0.583919 |
2
|
4734
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0009058
|
biosynthetic process
|
0.642905 |
0.642905 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.643595 |
0.643595 |
1
|
2093
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.646807 |
0.646807 |
1
|
1623
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0016070
|
RNA metabolic process
|
0.656488 |
0.656488 |
1
|
1191
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.658716 |
0.658716 |
1
|
1789
|
|
GO:0008380
|
RNA splicing
|
0.669082 |
0.669082 |
1
|
277
|
|
GO:0044444
|
cytoplasmic part
|
0.669378 |
0.669378 |
1
|
1863
|
|
GO:0044238
|
primary metabolic process
|
0.672346 |
0.672346 |
2
|
4259
|
|
GO:0009987
|
cellular process
|
0.673638 |
0.673638 |
2
|
6560
|
|
GO:0032501
|
multicellular organismal process
|
0.674757 |
0.674757 |
1
|
3315
|
|
GO:0071013
|
catalytic step 2 spliceosome
|
0.683333 |
0.683333 |
1
|
123
|
|
GO:0044267
|
cellular protein metabolic process
|
0.686538 |
0.686538 |
1
|
1505
|
|
GO:0003676
|
nucleic acid binding
|
0.697754 |
0.697754 |
1
|
1855
|
|
GO:0003743
|
translation initiation factor activity
|
0.698795 |
0.698795 |
1
|
58
|
|
GO:0007606
|
sensory perception of chemical stimulus
|
0.707746 |
0.707746 |
1
|
201
|
|
GO:0019538
|
protein metabolic process
|
0.724907 |
0.724907 |
1
|
2053
|
|
GO:0006397
|
mRNA processing
|
0.734411 |
0.734411 |
1
|
318
|
|
GO:0010468
|
regulation of gene expression
|
0.739954 |
0.739954 |
1
|
973
|
|
GO:0044446
|
intracellular organelle part
|
0.743053 |
0.743053 |
1
|
2162
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.746994 |
0.746994 |
1
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.748407 |
0.748407 |
1
|
1959
|
|
GO:0044249
|
cellular biosynthetic process
|
0.776153 |
0.776153 |
1
|
2034
|
|
GO:0043226
|
organelle
|
0.785296 |
0.785296 |
1
|
3537
|
|
GO:0005623
|
cell
|
0.787322 |
0.787322 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.787322 |
0.787322 |
2
|
6195
|
|
GO:0005737
|
cytoplasm
|
0.790684 |
0.790684 |
1
|
2378
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0071011
|
precatalytic spliceosome
|
0.822222 |
0.822222 |
1
|
148
|
|
GO:0000398
|
nuclear mRNA splicing, via spliceosome
|
0.836478 |
0.836478 |
1
|
266
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0000381
|
regulation of alternative nuclear mRNA splicing, via spliceosome
|
0.890625 |
0.890625 |
1
|
57
|
|
GO:0043234
|
protein complex
|
0.925620 |
0.925620 |
1
|
1564
|
|
GO:0050877
|
neurological system process
|
0.953313 |
0.953313 |
1
|
633
|
|
GO:0043229
|
intracellular organelle
|
0.961659 |
0.961659 |
1
|
3530
|
|
GO:0000375
|
RNA splicing, via transesterification reactions
|
0.963899 |
0.963899 |
1
|
267
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0000377
|
RNA splicing, via transesterification reactions with bulged adenosine as nucleophile
|
0.996255 |
0.996255 |
1
|
266
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|