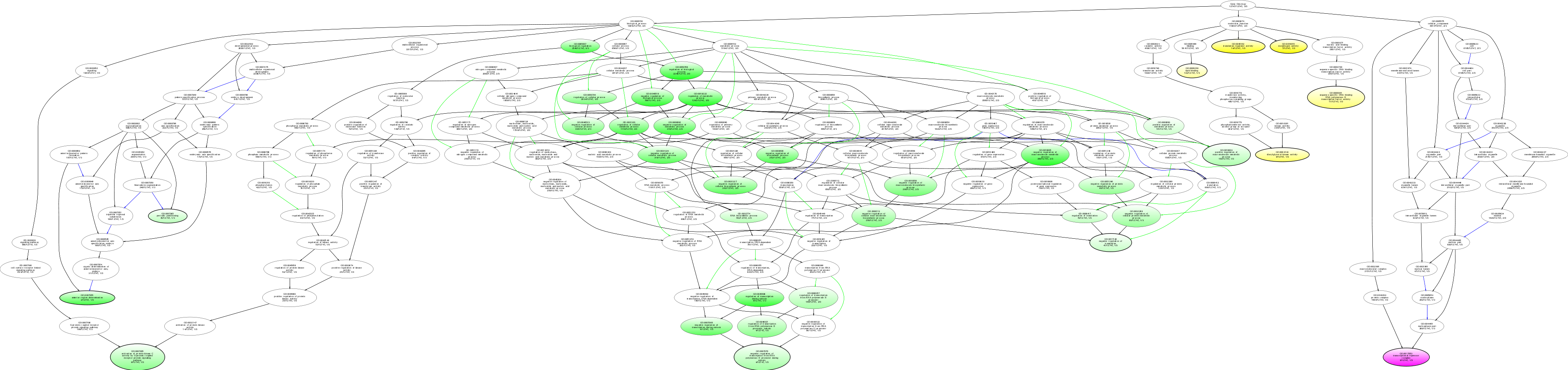
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0016015
|
morphogen activity
|
0.00189702 |
0.00189702 |
1
|
7
|
|
GO:0045182
|
translation regulator activity
|
0.00379164 |
0.00379164 |
1
|
14
|
|
GO:0017053
|
transcriptional repressor complex
|
0.00557967 |
0.00557967 |
1
|
9
|
|
GO:0019222
|
regulation of metabolic process
|
0.00929646 |
0.00929646 |
3
|
1242
|
|
GO:0050789
|
regulation of biological process
|
0.00945995 |
0.00945995 |
3
|
2246
|
|
GO:0010605
|
negative regulation of macromolecule metabolic process
|
0.00974342 |
0.00974342 |
2
|
356
|
|
GO:0048519
|
negative regulation of biological process
|
0.0126646 |
0.0126646 |
2
|
706
|
|
GO:0045896
|
regulation of transcription during mitosis
|
0.0128720 |
0.0128720 |
1
|
4
|
|
GO:0009892
|
negative regulation of metabolic process
|
0.0136708 |
0.0136708 |
2
|
372
|
|
GO:0065007
|
biological regulation
|
0.0138142 |
0.0138142 |
3
|
2548
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.0146239 |
0.0146239 |
3
|
1110
|
|
GO:0007355
|
anterior region determination
|
0.0152284 |
0.0152284 |
1
|
3
|
|
GO:0009890
|
negative regulation of biosynthetic process
|
0.0167740 |
0.0167740 |
2
|
282
|
|
GO:0031324
|
negative regulation of cellular metabolic process
|
0.0178283 |
0.0178283 |
2
|
319
|
|
GO:0031327
|
negative regulation of cellular biosynthetic process
|
0.0184844 |
0.0184844 |
2
|
282
|
|
GO:0007068
|
negative regulation of transcription during mitosis
|
0.0205128 |
0.0205128 |
1
|
4
|
|
GO:0004143
|
diacylglycerol kinase activity
|
0.0226131 |
0.0226131 |
1
|
9
|
|
GO:0048523
|
negative regulation of cellular process
|
0.0261866 |
0.0261866 |
2
|
635
|
|
GO:0010558
|
negative regulation of macromolecule biosynthetic process
|
0.0271748 |
0.0271748 |
2
|
280
|
|
GO:0000981
|
sequence-specific DNA binding RNA polymerase II transcription factor activity
|
0.0278481 |
0.0278481 |
1
|
11
|
|
GO:0050794
|
regulation of cellular process
|
0.0287277 |
0.0287277 |
3
|
2034
|
|
GO:2000113
|
negative regulation of cellular macromolecule biosynthetic process
|
0.0298220 |
0.0298220 |
2
|
280
|
|
GO:0046021
|
regulation of transcription from RNA polymerase II promoter, mitotic
|
0.0419903 |
0.0419903 |
1
|
4
|
|
GO:0007205
|
activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway
|
0.0441176 |
0.0441176 |
1
|
9
|
|
GO:0051248
|
negative regulation of protein metabolic process
|
0.0509361 |
0.0509361 |
1
|
60
|
|
GO:0032269
|
negative regulation of cellular protein metabolic process
|
0.0676809 |
0.0676809 |
1
|
60
|
|
GO:0008289
|
lipid binding
|
0.0690895 |
0.0690895 |
1
|
133
|
|
GO:0007070
|
negative regulation of transcription from RNA polymerase II promoter during mitosis
|
0.0714286 |
0.0714286 |
1
|
4
|
|
GO:0006357
|
regulation of transcription from RNA polymerase II promoter
|
0.0797476 |
0.0797476 |
2
|
189
|
|
GO:0010604
|
positive regulation of macromolecule metabolic process
|
0.0814533 |
0.0814533 |
1
|
150
|
|
GO:0032774
|
RNA biosynthetic process
|
0.0840246 |
0.0840246 |
2
|
703
|
|
GO:0007365
|
periodic partitioning
|
0.0889328 |
0.0889328 |
1
|
45
|
|
GO:0006417
|
regulation of translation
|
0.0934356 |
0.0934356 |
1
|
70
|
|
GO:0017148
|
negative regulation of translation
|
0.0936604 |
0.0936604 |
1
|
41
|
|
GO:0009893
|
positive regulation of metabolic process
|
0.0989758 |
0.0989758 |
1
|
181
|
|
GO:0051246
|
regulation of protein metabolic process
|
0.102375 |
0.102375 |
1
|
155
|
|
GO:0001071
|
nucleic acid binding transcription factor activity
|
0.103335 |
0.103335 |
1
|
395
|
|
GO:0051173
|
positive regulation of nitrogen compound metabolic process
|
0.108443 |
0.108443 |
1
|
119
|
|
GO:0048518
|
positive regulation of biological process
|
0.111456 |
0.111456 |
1
|
410
|
|
GO:0030528
|
transcription regulator activity
|
0.112632 |
0.112632 |
1
|
432
|
|
GO:0008152
|
metabolic process
|
0.117083 |
0.117083 |
3
|
5194
|
|
GO:0044237
|
cellular metabolic process
|
0.118315 |
0.118315 |
3
|
3814
|
|
GO:0010628
|
positive regulation of gene expression
|
0.123263 |
0.123263 |
1
|
122
|
|
GO:0045935
|
positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.125793 |
0.125793 |
1
|
119
|
|
GO:0031325
|
positive regulation of cellular metabolic process
|
0.125972 |
0.125972 |
1
|
172
|
|
GO:0006366
|
transcription from RNA polymerase II promoter
|
0.129929 |
0.129929 |
2
|
253
|
|
GO:0051254
|
positive regulation of RNA metabolic process
|
0.130868 |
0.130868 |
1
|
83
|
|
GO:0032583
|
regulation of gene-specific transcription
|
0.130997 |
0.130997 |
1
|
42
|
|
GO:0005488
|
binding
|
0.132501 |
0.132501 |
3
|
5641
|
|
GO:0019220
|
regulation of phosphate metabolic process
|
0.136691 |
0.136691 |
1
|
76
|
|
GO:0051174
|
regulation of phosphorus metabolic process
|
0.138139 |
0.138139 |
1
|
76
|
|
GO:0031974
|
membrane-enclosed lumen
|
0.141385 |
0.141385 |
1
|
647
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.142051 |
0.142051 |
2
|
1039
|
|
GO:0009891
|
positive regulation of biosynthetic process
|
0.146409 |
0.146409 |
1
|
160
|
|
GO:0031328
|
positive regulation of cellular biosynthetic process
|
0.149561 |
0.149561 |
1
|
159
|
|
GO:0032268
|
regulation of cellular protein metabolic process
|
0.149672 |
0.149672 |
1
|
128
|
|
GO:0006350
|
transcription
|
0.150290 |
0.150290 |
2
|
859
|
|
GO:0010557
|
positive regulation of macromolecule biosynthetic process
|
0.151735 |
0.151735 |
1
|
131
|
|
GO:0048522
|
positive regulation of cellular process
|
0.160427 |
0.160427 |
1
|
372
|
|
GO:0042325
|
regulation of phosphorylation
|
0.160839 |
0.160839 |
1
|
69
|
|
GO:0043226
|
organelle
|
0.160900 |
0.160900 |
2
|
3537
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.164491 |
0.164491 |
2
|
1623
|
|
GO:0016070
|
RNA metabolic process
|
0.171170 |
0.171170 |
2
|
1191
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.172797 |
0.172797 |
2
|
1789
|
|
GO:0065009
|
regulation of molecular function
|
0.184087 |
0.184087 |
1
|
167
|
|
GO:0007354
|
zygotic determination of anterior/posterior axis, embryo
|
0.184524 |
0.184524 |
1
|
31
|
|
GO:0043193
|
positive regulation of gene-specific transcription
|
0.186275 |
0.186275 |
1
|
19
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.193713 |
0.193713 |
2
|
686
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.194907 |
0.194907 |
2
|
1056
|
|
GO:0032569
|
gene-specific transcription from RNA polymerase II promoter
|
0.195338 |
0.195338 |
1
|
26
|
|
GO:0019992
|
diacylglycerol binding
|
0.195489 |
0.195489 |
1
|
26
|
|
GO:0023046
|
signaling process
|
0.199779 |
0.199779 |
1
|
760
|
|
GO:0007389
|
pattern specification process
|
0.203563 |
0.203563 |
1
|
537
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.208706 |
0.208706 |
2
|
850
|
|
GO:0045893
|
positive regulation of transcription, DNA-dependent
|
0.216360 |
0.216360 |
1
|
82
|
|
GO:0051172
|
negative regulation of nitrogen compound metabolic process
|
0.216720 |
0.216720 |
1
|
250
|
|
GO:0009987
|
cellular process
|
0.235914 |
0.235914 |
3
|
6560
|
|
GO:0045941
|
positive regulation of transcription
|
0.239105 |
0.239105 |
1
|
115
|
|
GO:0010468
|
regulation of gene expression
|
0.239905 |
0.239905 |
2
|
973
|
|
GO:0009790
|
embryo development
|
0.242987 |
0.242987 |
1
|
641
|
|
GO:0010551
|
regulation of gene-specific transcription from RNA polymerase II promoter
|
0.243668 |
0.243668 |
1
|
26
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.246859 |
0.246859 |
2
|
1617
|
|
GO:0010608
|
posttranscriptional regulation of gene expression
|
0.249483 |
0.249483 |
1
|
130
|
|
GO:0016740
|
transferase activity
|
0.250852 |
0.250852 |
1
|
1030
|
|
GO:0045934
|
negative regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.252452 |
0.252452 |
1
|
250
|
|
GO:0000003
|
reproduction
|
0.261920 |
0.261920 |
1
|
1022
|
|
GO:0022414
|
reproductive process
|
0.261920 |
0.261920 |
1
|
1022
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.263629 |
0.263629 |
2
|
850
|
|
GO:0000122
|
negative regulation of transcription from RNA polymerase II promoter
|
0.270076 |
0.270076 |
1
|
56
|
|
GO:0051347
|
positive regulation of transferase activity
|
0.271739 |
0.271739 |
1
|
25
|
|
GO:0010629
|
negative regulation of gene expression
|
0.276112 |
0.276112 |
1
|
288
|
|
GO:0048609
|
multicellular organismal reproductive process
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0032504
|
multicellular organism reproduction
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0010467
|
gene expression
|
0.282416 |
0.282416 |
2
|
1907
|
|
GO:0051253
|
negative regulation of RNA metabolic process
|
0.287333 |
0.287333 |
1
|
202
|
|
GO:0010552
|
positive regulation of gene-specific transcription from RNA polymerase II promoter
|
0.288136 |
0.288136 |
1
|
17
|
|
GO:0043233
|
organelle lumen
|
0.291360 |
0.291360 |
1
|
634
|
|
GO:0003704
|
specific RNA polymerase II transcription factor activity
|
0.292135 |
0.292135 |
1
|
78
|
|
GO:0070013
|
intracellular organelle lumen
|
0.293247 |
0.293247 |
1
|
634
|
|
GO:0009880
|
embryonic pattern specification
|
0.294591 |
0.294591 |
1
|
256
|
|
GO:0003729
|
mRNA binding
|
0.302147 |
0.302147 |
1
|
197
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.304837 |
0.304837 |
2
|
888
|
|
GO:0045944
|
positive regulation of transcription from RNA polymerase II promoter
|
0.311956 |
0.311956 |
1
|
48
|
|
GO:0044451
|
nucleoplasm part
|
0.313253 |
0.313253 |
1
|
260
|
|
GO:0050790
|
regulation of catalytic activity
|
0.317558 |
0.317558 |
1
|
149
|
|
GO:0007186
|
G-protein coupled receptor protein signaling pathway
|
0.318910 |
0.318910 |
1
|
199
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.325784 |
0.325784 |
2
|
905
|
|
GO:0007351
|
tripartite regional subdivision
|
0.332016 |
0.332016 |
1
|
168
|
|
GO:0023052
|
signaling
|
0.338079 |
0.338079 |
1
|
1364
|
|
GO:0005654
|
nucleoplasm
|
0.340964 |
0.340964 |
1
|
283
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.346544 |
0.346544 |
2
|
888
|
|
GO:0003723
|
RNA binding
|
0.351482 |
0.351482 |
1
|
652
|
|
GO:0009058
|
biosynthetic process
|
0.355414 |
0.355414 |
2
|
2090
|
|
GO:0051338
|
regulation of transferase activity
|
0.355705 |
0.355705 |
1
|
53
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.356248 |
0.356248 |
2
|
2093
|
|
GO:0006793
|
phosphorus metabolic process
|
0.376764 |
0.376764 |
1
|
556
|
|
GO:0006412
|
translation
|
0.382593 |
0.382593 |
1
|
600
|
|
GO:0009952
|
anterior/posterior pattern formation
|
0.389328 |
0.389328 |
1
|
197
|
|
GO:0043085
|
positive regulation of catalytic activity
|
0.405063 |
0.405063 |
1
|
64
|
|
GO:0005634
|
nucleus
|
0.408695 |
0.408695 |
2
|
1826
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.427778 |
0.427778 |
2
|
903
|
|
GO:0016481
|
negative regulation of transcription
|
0.428239 |
0.428239 |
1
|
233
|
|
GO:0032991
|
macromolecular complex
|
0.428425 |
0.428425 |
1
|
2151
|
|
GO:0023034
|
intracellular signaling pathway
|
0.430962 |
0.430962 |
1
|
412
|
|
GO:0009798
|
axis specification
|
0.432030 |
0.432030 |
1
|
232
|
|
GO:0044422
|
organelle part
|
0.432705 |
0.432705 |
1
|
2176
|
|
GO:0044093
|
positive regulation of molecular function
|
0.437126 |
0.437126 |
1
|
73
|
|
GO:0045892
|
negative regulation of transcription, DNA-dependent
|
0.461555 |
0.461555 |
1
|
195
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.468064 |
0.468064 |
2
|
620
|
|
GO:0044428
|
nuclear part
|
0.469235 |
0.469235 |
1
|
830
|
|
GO:0033674
|
positive regulation of kinase activity
|
0.471698 |
0.471698 |
1
|
25
|
|
GO:0045860
|
positive regulation of protein kinase activity
|
0.471698 |
0.471698 |
1
|
25
|
|
GO:0045449
|
regulation of transcription
|
0.472005 |
0.472005 |
2
|
771
|
|
GO:0031981
|
nuclear lumen
|
0.475277 |
0.475277 |
1
|
471
|
|
GO:0016772
|
transferase activity, transferring phosphorus-containing groups
|
0.480583 |
0.480583 |
1
|
495
|
|
GO:0005623
|
cell
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.497519 |
0.497519 |
2
|
1959
|
|
GO:0044424
|
intracellular part
|
0.500760 |
0.500760 |
2
|
4384
|
|
GO:0044249
|
cellular biosynthetic process
|
0.540182 |
0.540182 |
2
|
2034
|
|
GO:0000578
|
embryonic axis specification
|
0.552716 |
0.552716 |
1
|
173
|
|
GO:0032147
|
activation of protein kinase activity
|
0.560000 |
0.560000 |
1
|
14
|
|
GO:0032502
|
developmental process
|
0.575616 |
0.575616 |
1
|
2638
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0035282
|
segmentation
|
0.581028 |
0.581028 |
1
|
294
|
|
GO:0005622
|
intracellular
|
0.583919 |
0.583919 |
2
|
4734
|
|
GO:0007165
|
signal transduction
|
0.585130 |
0.585130 |
1
|
548
|
|
GO:0008358
|
maternal determination of anterior/posterior axis, embryo
|
0.613095 |
0.613095 |
1
|
103
|
|
GO:0003702
|
RNA polymerase II transcription factor activity
|
0.618056 |
0.618056 |
1
|
267
|
|
GO:0043229
|
intracellular organelle
|
0.646541 |
0.646541 |
2
|
3530
|
|
GO:0007166
|
cell surface receptor linked signaling pathway
|
0.652720 |
0.652720 |
1
|
624
|
|
GO:0043227
|
membrane-bounded organelle
|
0.653325 |
0.653325 |
2
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.653801 |
0.653801 |
2
|
2856
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.662696 |
0.662696 |
2
|
701
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0005515
|
protein binding
|
0.665786 |
0.665786 |
1
|
1726
|
|
GO:0032501
|
multicellular organismal process
|
0.674757 |
0.674757 |
1
|
3315
|
|
GO:0009948
|
anterior/posterior axis specification
|
0.675781 |
0.675781 |
1
|
173
|
|
GO:0044267
|
cellular protein metabolic process
|
0.686538 |
0.686538 |
1
|
1505
|
|
GO:0007367
|
segment polarity determination
|
0.688889 |
0.688889 |
1
|
31
|
|
GO:0003676
|
nucleic acid binding
|
0.697754 |
0.697754 |
1
|
1855
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.703351 |
0.703351 |
2
|
2878
|
|
GO:0019538
|
protein metabolic process
|
0.724907 |
0.724907 |
1
|
2053
|
|
GO:0043234
|
protein complex
|
0.727104 |
0.727104 |
1
|
1564
|
|
GO:0007292
|
female gamete generation
|
0.727380 |
0.727380 |
1
|
619
|
|
GO:0016773
|
phosphotransferase activity, alcohol group as acceptor
|
0.735354 |
0.735354 |
1
|
364
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.736132 |
0.736132 |
2
|
1535
|
|
GO:0044446
|
intracellular organelle part
|
0.743053 |
0.743053 |
1
|
2162
|
|
GO:0003824
|
catalytic activity
|
0.751316 |
0.751316 |
1
|
4106
|
|
GO:0016310
|
phosphorylation
|
0.758993 |
0.758993 |
1
|
422
|
|
GO:0043549
|
regulation of kinase activity
|
0.768116 |
0.768116 |
1
|
53
|
|
GO:0043170
|
macromolecule metabolic process
|
0.772351 |
0.772351 |
2
|
3588
|
|
GO:0005575
|
cellular_component
|
0.773470 |
0.773470 |
2
|
8817
|
|
GO:0007350
|
blastoderm segmentation
|
0.779221 |
0.779221 |
1
|
240
|
|
GO:0016301
|
kinase activity
|
0.795960 |
0.795960 |
1
|
394
|
|
GO:0019953
|
sexual reproduction
|
0.859100 |
0.859100 |
1
|
878
|
|
GO:0007276
|
gamete generation
|
0.883697 |
0.883697 |
1
|
851
|
|
GO:0044238
|
primary metabolic process
|
0.914505 |
0.914505 |
2
|
4259
|
|
GO:0003002
|
regionalization
|
0.942272 |
0.942272 |
1
|
506
|
|
GO:0008595
|
anterior/posterior axis specification, embryo
|
0.943820 |
0.943820 |
1
|
168
|
|
GO:0048477
|
oogenesis
|
0.987076 |
0.987076 |
1
|
611
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0003705
|
sequence-specific enhancer binding RNA polymerase II transcription factor activity
|
1.00000 |
1.00000 |
1
|
11
|
|
GO:0003700
|
sequence-specific DNA binding transcription factor activity
|
1.00000 |
1.00000 |
1
|
395
|
|
GO:0006796
|
phosphate metabolic process
|
1.00000 |
1.00000 |
1
|
556
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|
|
GO:0045859
|
regulation of protein kinase activity
|
1.00000 |
1.00000 |
1
|
53
|