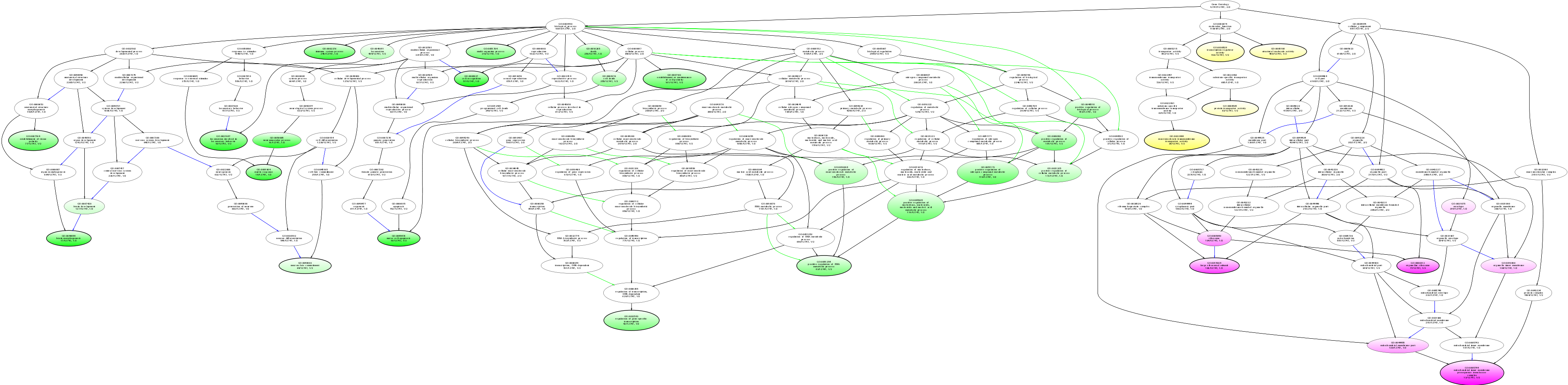
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0005744
|
mitochondrial inner membrane presequence translocase complex
|
0.00798526 |
0.00798526 |
1
|
13
|
|
GO:0008037
|
cell recognition
|
0.0147896 |
0.0147896 |
1
|
57
|
|
GO:0045476
|
nurse cell apoptosis
|
0.0176991 |
0.0176991 |
1
|
16
|
|
GO:0048854
|
brain morphogenesis
|
0.0221354 |
0.0221354 |
1
|
17
|
|
GO:0050905
|
neuromuscular process
|
0.0252765 |
0.0252765 |
1
|
16
|
|
GO:0031987
|
locomotion involved in locomotory behavior
|
0.0266667 |
0.0266667 |
1
|
16
|
|
GO:0000313
|
organellar ribosome
|
0.0350957 |
0.0350957 |
1
|
77
|
|
GO:0001964
|
startle response
|
0.0400000 |
0.0400000 |
1
|
15
|
|
GO:0002376
|
immune system process
|
0.0456263 |
0.0456263 |
1
|
245
|
|
GO:0015934
|
large ribosomal subunit
|
0.0477693 |
0.0477693 |
1
|
106
|
|
GO:0016265
|
death
|
0.0482013 |
0.0482013 |
1
|
259
|
|
GO:0007163
|
establishment or maintenance of cell polarity
|
0.0484867 |
0.0484867 |
1
|
161
|
|
GO:0007164
|
establishment of tissue polarity
|
0.0501956 |
0.0501956 |
1
|
77
|
|
GO:0051173
|
positive regulation of nitrogen compound metabolic process
|
0.0557638 |
0.0557638 |
1
|
119
|
|
GO:0022884
|
macromolecule transmembrane transporter activity
|
0.0560000 |
0.0560000 |
1
|
35
|
|
GO:0051704
|
multi-organism process
|
0.0591978 |
0.0591978 |
1
|
319
|
|
GO:0045935
|
positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.0649918 |
0.0649918 |
1
|
119
|
|
GO:0009893
|
positive regulation of metabolic process
|
0.0671169 |
0.0671169 |
1
|
181
|
|
GO:0051254
|
positive regulation of RNA metabolic process
|
0.0676998 |
0.0676998 |
1
|
83
|
|
GO:0032583
|
regulation of gene-specific transcription
|
0.0677419 |
0.0677419 |
1
|
42
|
|
GO:0005840
|
ribosome
|
0.0681419 |
0.0681419 |
1
|
194
|
|
GO:0048518
|
positive regulation of biological process
|
0.0757538 |
0.0757538 |
1
|
410
|
|
GO:0008219
|
cell death
|
0.0762386 |
0.0762386 |
1
|
255
|
|
GO:0030528
|
transcription regulator activity
|
0.0765699 |
0.0765699 |
1
|
432
|
|
GO:0044455
|
mitochondrial membrane part
|
0.0795594 |
0.0795594 |
1
|
130
|
|
GO:0005198
|
structural molecule activity
|
0.0812543 |
0.0812543 |
1
|
459
|
|
GO:0010604
|
positive regulation of macromolecule metabolic process
|
0.0814533 |
0.0814533 |
1
|
150
|
|
GO:0031325
|
positive regulation of cellular metabolic process
|
0.0858406 |
0.0858406 |
1
|
172
|
|
GO:0019866
|
organelle inner membrane
|
0.0865358 |
0.0865358 |
1
|
194
|
|
GO:0040011
|
locomotion
|
0.0891086 |
0.0891086 |
1
|
484
|
|
GO:0008565
|
protein transporter activity
|
0.0906475 |
0.0906475 |
1
|
63
|
|
GO:0048663
|
neuron fate commitment
|
0.0942563 |
0.0942563 |
1
|
64
|
|
GO:0031975
|
envelope
|
0.0988108 |
0.0988108 |
1
|
314
|
|
GO:0007420
|
brain development
|
0.0990640 |
0.0990640 |
1
|
127
|
|
GO:0009607
|
response to biotic stimulus
|
0.103359 |
0.103359 |
1
|
160
|
|
GO:0048522
|
positive regulation of cellular process
|
0.110028 |
0.110028 |
1
|
372
|
|
GO:0042067
|
establishment of ommatidial planar polarity
|
0.111814 |
0.111814 |
1
|
53
|
|
GO:0048056
|
R3/R4 cell differentiation
|
0.111940 |
0.111940 |
1
|
15
|
|
GO:0006955
|
immune response
|
0.113493 |
0.113493 |
1
|
180
|
|
GO:0045893
|
positive regulation of transcription, DNA-dependent
|
0.114685 |
0.114685 |
1
|
82
|
|
GO:0008038
|
neuron recognition
|
0.118644 |
0.118644 |
1
|
56
|
|
GO:0016198
|
axon choice point recognition
|
0.120000 |
0.120000 |
1
|
24
|
|
GO:0010628
|
positive regulation of gene expression
|
0.123263 |
0.123263 |
1
|
122
|
|
GO:0005762
|
mitochondrial large ribosomal subunit
|
0.125000 |
0.125000 |
1
|
48
|
|
GO:0044429
|
mitochondrial part
|
0.125285 |
0.125285 |
1
|
384
|
|
GO:0045941
|
positive regulation of transcription
|
0.127636 |
0.127636 |
1
|
115
|
|
GO:0007417
|
central nervous system development
|
0.135613 |
0.135613 |
1
|
230
|
|
GO:0015450
|
P-P-bond-hydrolysis-driven protein transmembrane transporter activity
|
0.137931 |
0.137931 |
1
|
24
|
|
GO:0016563
|
transcription activator activity
|
0.141204 |
0.141204 |
1
|
61
|
|
GO:0031974
|
membrane-enclosed lumen
|
0.141385 |
0.141385 |
1
|
647
|
|
GO:0030030
|
cell projection organization
|
0.142264 |
0.142264 |
1
|
485
|
|
GO:0009891
|
positive regulation of biosynthetic process
|
0.146409 |
0.146409 |
1
|
160
|
|
GO:0042461
|
photoreceptor cell development
|
0.147601 |
0.147601 |
1
|
80
|
|
GO:0005215
|
transporter activity
|
0.148089 |
0.148089 |
1
|
852
|
|
GO:0045467
|
R7 cell development
|
0.148936 |
0.148936 |
1
|
14
|
|
GO:0031328
|
positive regulation of cellular biosynthetic process
|
0.149561 |
0.149561 |
1
|
159
|
|
GO:0010557
|
positive regulation of macromolecule biosynthetic process
|
0.151735 |
0.151735 |
1
|
131
|
|
GO:0031090
|
organelle membrane
|
0.153645 |
0.153645 |
1
|
396
|
|
GO:0007464
|
R3/R4 cell fate commitment
|
0.153846 |
0.153846 |
1
|
14
|
|
GO:0043226
|
organelle
|
0.160900 |
0.160900 |
2
|
3537
|
|
GO:0009058
|
biosynthetic process
|
0.161869 |
0.161869 |
2
|
2090
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.164491 |
0.164491 |
2
|
1623
|
|
GO:0045184
|
establishment of protein localization
|
0.168623 |
0.168623 |
1
|
273
|
|
GO:0031967
|
organelle envelope
|
0.169005 |
0.169005 |
1
|
304
|
|
GO:0051641
|
cellular localization
|
0.169056 |
0.169056 |
1
|
607
|
|
GO:0048610
|
cellular process involved in reproduction
|
0.173346 |
0.173346 |
1
|
613
|
|
GO:0015031
|
protein transport
|
0.177333 |
0.177333 |
1
|
266
|
|
GO:0005759
|
mitochondrial matrix
|
0.179606 |
0.179606 |
1
|
155
|
|
GO:0000003
|
reproduction
|
0.183280 |
0.183280 |
1
|
1022
|
|
GO:0022414
|
reproductive process
|
0.183280 |
0.183280 |
1
|
1022
|
|
GO:0032989
|
cellular component morphogenesis
|
0.193108 |
0.193108 |
1
|
566
|
|
GO:0048729
|
tissue morphogenesis
|
0.194151 |
0.194151 |
1
|
312
|
|
GO:0045165
|
cell fate commitment
|
0.199843 |
0.199843 |
1
|
255
|
|
GO:0003008
|
system process
|
0.200302 |
0.200302 |
1
|
664
|
|
GO:0005761
|
mitochondrial ribosome
|
0.200521 |
0.200521 |
1
|
77
|
|
GO:0051705
|
behavioral interaction between organisms
|
0.209431 |
0.209431 |
1
|
151
|
|
GO:0030529
|
ribonucleoprotein complex
|
0.210589 |
0.210589 |
1
|
510
|
|
GO:0046907
|
intracellular transport
|
0.227832 |
0.227832 |
1
|
352
|
|
GO:0008152
|
metabolic process
|
0.239353 |
0.239353 |
2
|
5194
|
|
GO:0044237
|
cellular metabolic process
|
0.241038 |
0.241038 |
2
|
3814
|
|
GO:0009605
|
response to external stimulus
|
0.242248 |
0.242248 |
1
|
375
|
|
GO:0009888
|
tissue development
|
0.245916 |
0.245916 |
1
|
557
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.246859 |
0.246859 |
2
|
1617
|
|
GO:0043167
|
ion binding
|
0.265556 |
0.265556 |
1
|
1498
|
|
GO:0050896
|
response to stimulus
|
0.270384 |
0.270384 |
1
|
1548
|
|
GO:0051234
|
establishment of localization
|
0.270545 |
0.270545 |
1
|
1549
|
|
GO:0033036
|
macromolecule localization
|
0.276371 |
0.276371 |
1
|
524
|
|
GO:0044249
|
cellular biosynthetic process
|
0.277461 |
0.277461 |
2
|
2034
|
|
GO:0048609
|
multicellular organismal reproductive process
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0032504
|
multicellular organism reproduction
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0007626
|
locomotory behavior
|
0.280859 |
0.280859 |
1
|
157
|
|
GO:0010467
|
gene expression
|
0.282416 |
0.282416 |
2
|
1907
|
|
GO:0006357
|
regulation of transcription from RNA polymerase II promoter
|
0.282934 |
0.282934 |
1
|
189
|
|
GO:0005739
|
mitochondrion
|
0.284169 |
0.284169 |
1
|
551
|
|
GO:0006935
|
chemotaxis
|
0.287630 |
0.287630 |
1
|
193
|
|
GO:0043233
|
organelle lumen
|
0.291360 |
0.291360 |
1
|
634
|
|
GO:0003704
|
specific RNA polymerase II transcription factor activity
|
0.292135 |
0.292135 |
1
|
78
|
|
GO:0070013
|
intracellular organelle lumen
|
0.293247 |
0.293247 |
1
|
634
|
|
GO:0046530
|
photoreceptor cell differentiation
|
0.295620 |
0.295620 |
1
|
162
|
|
GO:0051649
|
establishment of localization in cell
|
0.303951 |
0.303951 |
1
|
500
|
|
GO:0001738
|
morphogenesis of a polarized epithelium
|
0.304498 |
0.304498 |
1
|
88
|
|
GO:0005515
|
protein binding
|
0.305974 |
0.305974 |
1
|
1726
|
|
GO:0045466
|
R7 cell differentiation
|
0.313433 |
0.313433 |
1
|
42
|
|
GO:0048869
|
cellular developmental process
|
0.322329 |
0.322329 |
1
|
1276
|
|
GO:0051179
|
localization
|
0.325313 |
0.325313 |
1
|
1896
|
|
GO:0003676
|
nucleic acid binding
|
0.328842 |
0.328842 |
1
|
1855
|
|
GO:0046552
|
photoreceptor cell fate commitment
|
0.345238 |
0.345238 |
1
|
58
|
|
GO:0001751
|
compound eye photoreceptor cell differentiation
|
0.354497 |
0.354497 |
1
|
134
|
|
GO:0000315
|
organellar large ribosomal subunit
|
0.355556 |
0.355556 |
1
|
48
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.357722 |
0.357722 |
1
|
2108
|
|
GO:0006366
|
transcription from RNA polymerase II promoter
|
0.360913 |
0.360913 |
1
|
253
|
|
GO:0007610
|
behavior
|
0.361111 |
0.361111 |
1
|
559
|
|
GO:0001754
|
eye photoreceptor cell differentiation
|
0.361386 |
0.361386 |
1
|
146
|
|
GO:0042706
|
eye photoreceptor cell fate commitment
|
0.371622 |
0.371622 |
1
|
55
|
|
GO:0009887
|
organ morphogenesis
|
0.374384 |
0.374384 |
1
|
684
|
|
GO:0019222
|
regulation of metabolic process
|
0.376562 |
0.376562 |
1
|
1242
|
|
GO:0000904
|
cell morphogenesis involved in differentiation
|
0.376731 |
0.376731 |
1
|
408
|
|
GO:0050789
|
regulation of biological process
|
0.378390 |
0.378390 |
1
|
2246
|
|
GO:0031966
|
mitochondrial membrane
|
0.380531 |
0.380531 |
1
|
215
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.381695 |
0.381695 |
1
|
905
|
|
GO:0009987
|
cellular process
|
0.381822 |
0.381822 |
2
|
6560
|
|
GO:0006412
|
translation
|
0.382593 |
0.382593 |
1
|
600
|
|
GO:0042330
|
taxis
|
0.389803 |
0.389803 |
1
|
237
|
|
GO:0002118
|
aggressive behavior
|
0.397351 |
0.397351 |
1
|
60
|
|
GO:0003735
|
structural constituent of ribosome
|
0.398693 |
0.398693 |
1
|
183
|
|
GO:0044425
|
membrane part
|
0.400435 |
0.400435 |
1
|
1398
|
|
GO:0042221
|
response to chemical stimulus
|
0.408269 |
0.408269 |
1
|
632
|
|
GO:0070727
|
cellular macromolecule localization
|
0.408300 |
0.408300 |
1
|
305
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.408435 |
0.408435 |
2
|
2878
|
|
GO:0001752
|
compound eye photoreceptor fate commitment
|
0.410448 |
0.410448 |
1
|
55
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.418200 |
0.418200 |
1
|
1039
|
|
GO:0065007
|
biological regulation
|
0.422440 |
0.422440 |
1
|
2548
|
|
GO:0032991
|
macromolecular complex
|
0.428425 |
0.428425 |
1
|
2151
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.429576 |
0.429576 |
1
|
1110
|
|
GO:0044422
|
organelle part
|
0.432705 |
0.432705 |
1
|
2176
|
|
GO:0048666
|
neuron development
|
0.433702 |
0.433702 |
1
|
471
|
|
GO:0032502
|
developmental process
|
0.435255 |
0.435255 |
1
|
2638
|
|
GO:0007423
|
sensory organ development
|
0.438003 |
0.438003 |
1
|
544
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.440308 |
0.440308 |
1
|
686
|
|
GO:0030182
|
neuron differentiation
|
0.442292 |
0.442292 |
1
|
548
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.451726 |
0.451726 |
1
|
903
|
|
GO:0022008
|
neurogenesis
|
0.452398 |
0.452398 |
1
|
632
|
|
GO:0015399
|
primary active transmembrane transporter activity
|
0.452778 |
0.452778 |
1
|
163
|
|
GO:0048468
|
cell development
|
0.457419 |
0.457419 |
1
|
1042
|
|
GO:0042462
|
eye photoreceptor cell development
|
0.458065 |
0.458065 |
1
|
71
|
|
GO:0051707
|
response to other organism
|
0.473846 |
0.473846 |
1
|
154
|
|
GO:0043170
|
macromolecule metabolic process
|
0.477159 |
0.477159 |
2
|
3588
|
|
GO:0005575
|
cellular_component
|
0.478421 |
0.478421 |
2
|
8817
|
|
GO:0034613
|
cellular protein localization
|
0.478936 |
0.478936 |
1
|
216
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.486014 |
0.486014 |
1
|
1056
|
|
GO:0005623
|
cell
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0022804
|
active transmembrane transporter activity
|
0.495868 |
0.495868 |
1
|
360
|
|
GO:0032774
|
RNA biosynthetic process
|
0.496008 |
0.496008 |
1
|
703
|
|
GO:0007399
|
nervous system development
|
0.500000 |
0.500000 |
1
|
848
|
|
GO:0044424
|
intracellular part
|
0.500760 |
0.500760 |
2
|
4384
|
|
GO:0006886
|
intracellular protein transport
|
0.504878 |
0.504878 |
1
|
207
|
|
GO:0042051
|
compound eye photoreceptor development
|
0.507353 |
0.507353 |
1
|
69
|
|
GO:0005740
|
mitochondrial envelope
|
0.512088 |
0.512088 |
1
|
233
|
|
GO:0050794
|
regulation of cellular process
|
0.518911 |
0.518911 |
1
|
2034
|
|
GO:0032501
|
multicellular organismal process
|
0.527040 |
0.527040 |
1
|
3315
|
|
GO:0060429
|
epithelium development
|
0.529623 |
0.529623 |
1
|
295
|
|
GO:0048592
|
eye morphogenesis
|
0.542069 |
0.542069 |
1
|
393
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0008320
|
protein transmembrane transporter activity
|
0.555556 |
0.555556 |
1
|
35
|
|
GO:0016020
|
membrane
|
0.567775 |
0.567775 |
1
|
2122
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.573530 |
0.573530 |
1
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.574428 |
0.574428 |
1
|
1227
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0005622
|
intracellular
|
0.583919 |
0.583919 |
2
|
4734
|
|
GO:0019730
|
antimicrobial humoral response
|
0.584337 |
0.584337 |
1
|
97
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.599142 |
0.599142 |
1
|
888
|
|
GO:0006959
|
humoral immune response
|
0.616667 |
0.616667 |
1
|
111
|
|
GO:0003702
|
RNA polymerase II transcription factor activity
|
0.618056 |
0.618056 |
1
|
267
|
|
GO:0006350
|
transcription
|
0.625331 |
0.625331 |
1
|
859
|
|
GO:0006915
|
apoptosis
|
0.633333 |
0.633333 |
1
|
152
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.635595 |
0.635595 |
1
|
888
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.643595 |
0.643595 |
1
|
2093
|
|
GO:0043229
|
intracellular organelle
|
0.646541 |
0.646541 |
2
|
3530
|
|
GO:0043227
|
membrane-bounded organelle
|
0.653325 |
0.653325 |
2
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.653801 |
0.653801 |
2
|
2856
|
|
GO:0016070
|
RNA metabolic process
|
0.656488 |
0.656488 |
1
|
1191
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.658716 |
0.658716 |
1
|
1789
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0044444
|
cytoplasmic part
|
0.669378 |
0.669378 |
1
|
1863
|
|
GO:0044238
|
primary metabolic process
|
0.672346 |
0.672346 |
2
|
4259
|
|
GO:0007409
|
axonogenesis
|
0.679389 |
0.679389 |
1
|
267
|
|
GO:0007411
|
axon guidance
|
0.683824 |
0.683824 |
1
|
186
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.684327 |
0.684327 |
1
|
620
|
|
GO:0044267
|
cellular protein metabolic process
|
0.686538 |
0.686538 |
1
|
1505
|
|
GO:0045449
|
regulation of transcription
|
0.687166 |
0.687166 |
1
|
771
|
|
GO:0008150
|
biological_process
|
0.693584 |
0.693584 |
2
|
10616
|
|
GO:0031175
|
neuron projection development
|
0.696270 |
0.696270 |
1
|
392
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.705273 |
0.705273 |
1
|
850
|
|
GO:0019538
|
protein metabolic process
|
0.724907 |
0.724907 |
1
|
2053
|
|
GO:0008104
|
protein localization
|
0.727099 |
0.727099 |
1
|
381
|
|
GO:0043234
|
protein complex
|
0.727104 |
0.727104 |
1
|
1564
|
|
GO:0007292
|
female gamete generation
|
0.727380 |
0.727380 |
1
|
619
|
|
GO:0010468
|
regulation of gene expression
|
0.739954 |
0.739954 |
1
|
973
|
|
GO:0044446
|
intracellular organelle part
|
0.743053 |
0.743053 |
1
|
2162
|
|
GO:0032990
|
cell part morphogenesis
|
0.745583 |
0.745583 |
1
|
422
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.748407 |
0.748407 |
1
|
1959
|
|
GO:0003674
|
molecular_function
|
0.753361 |
0.753361 |
2
|
11064
|
|
GO:0005488
|
binding
|
0.759777 |
0.759777 |
1
|
5641
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.763561 |
0.763561 |
1
|
850
|
|
GO:0008270
|
zinc ion binding
|
0.781440 |
0.781440 |
1
|
901
|
|
GO:0022891
|
substrate-specific transmembrane transporter activity
|
0.788146 |
0.788146 |
1
|
625
|
|
GO:0005737
|
cytoplasm
|
0.790684 |
0.790684 |
1
|
2378
|
|
GO:0016199
|
axon midline choice point recognition
|
0.791667 |
0.791667 |
1
|
19
|
|
GO:0048667
|
cell morphogenesis involved in neuron differentiation
|
0.795501 |
0.795501 |
1
|
389
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0048858
|
cell projection morphogenesis
|
0.806883 |
0.806883 |
1
|
422
|
|
GO:0001745
|
compound eye morphogenesis
|
0.811947 |
0.811947 |
1
|
367
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.814170 |
0.814170 |
1
|
701
|
|
GO:0022892
|
substrate-specific transporter activity
|
0.815728 |
0.815728 |
1
|
695
|
|
GO:0000902
|
cell morphogenesis
|
0.825088 |
0.825088 |
1
|
467
|
|
GO:0001654
|
eye development
|
0.841912 |
0.841912 |
1
|
458
|
|
GO:0005743
|
mitochondrial inner membrane
|
0.842342 |
0.842342 |
1
|
187
|
|
GO:0022857
|
transmembrane transporter activity
|
0.852113 |
0.852113 |
1
|
726
|
|
GO:0001736
|
establishment of planar polarity
|
0.853933 |
0.853933 |
1
|
76
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0019953
|
sexual reproduction
|
0.859100 |
0.859100 |
1
|
878
|
|
GO:0005634
|
nucleus
|
0.870016 |
0.870016 |
1
|
1826
|
|
GO:0007276
|
gamete generation
|
0.883697 |
0.883697 |
1
|
851
|
|
GO:0002009
|
morphogenesis of an epithelium
|
0.908805 |
0.908805 |
1
|
289
|
|
GO:0048812
|
neuron projection morphogenesis
|
0.910798 |
0.910798 |
1
|
388
|
|
GO:0048749
|
compound eye development
|
0.932314 |
0.932314 |
1
|
427
|
|
GO:0012501
|
programmed cell death
|
0.941176 |
0.941176 |
1
|
240
|
|
GO:0050877
|
neurological system process
|
0.953313 |
0.953313 |
1
|
633
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0002121
|
inter-male aggressive behavior
|
0.966667 |
0.966667 |
1
|
58
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0048477
|
oogenesis
|
0.987076 |
0.987076 |
1
|
611
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
2
|
12747
|
|
GO:0015405
|
P-P-bond-hydrolysis-driven transmembrane transporter activity
|
1.00000 |
1.00000 |
1
|
163
|