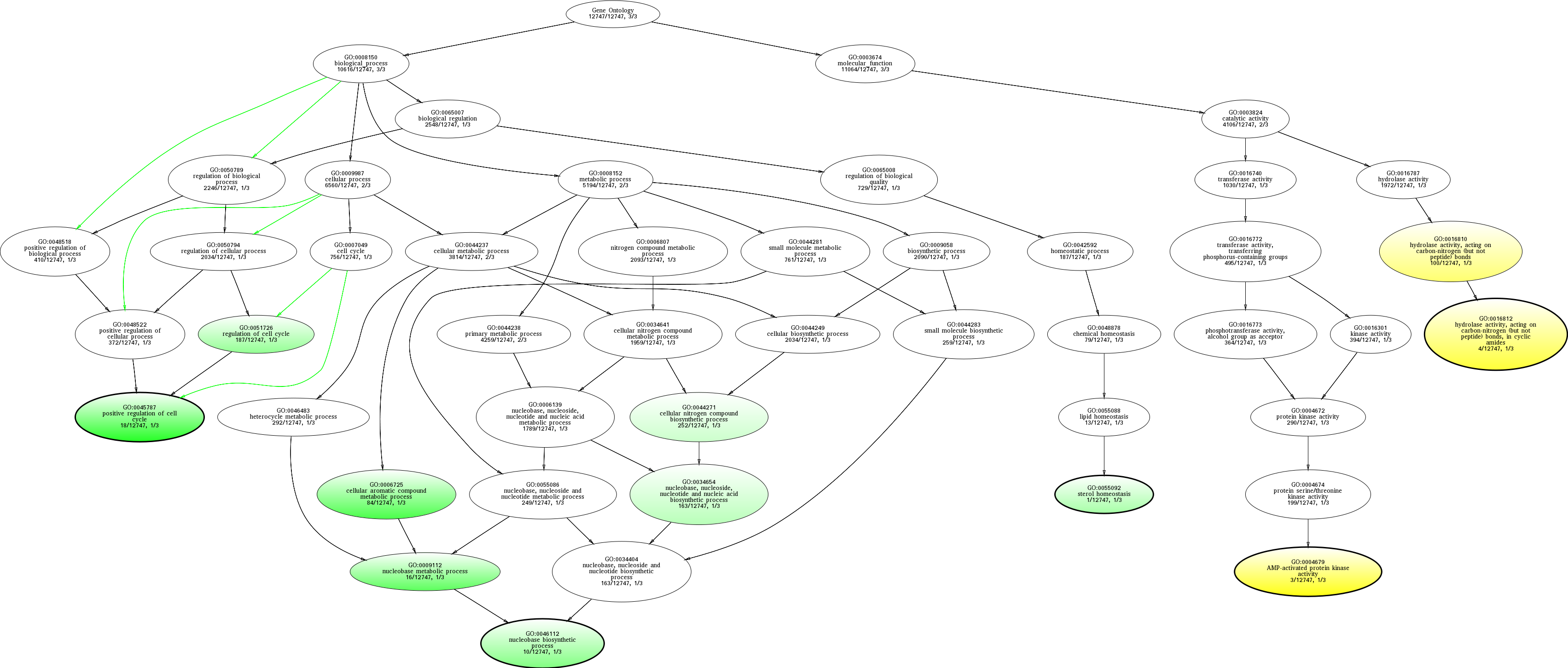
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0004679
|
AMP-activated protein kinase activity
|
0.0150754 |
0.0150754 |
1
|
3
|
|
GO:0045787
|
positive regulation of cell cycle
|
0.0171103 |
0.0171103 |
1
|
18
|
|
GO:0016812
|
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides
|
0.0400000 |
0.0400000 |
1
|
4
|
|
GO:0006725
|
cellular aromatic compound metabolic process
|
0.0435688 |
0.0435688 |
1
|
84
|
|
GO:0009112
|
nucleobase metabolic process
|
0.0441989 |
0.0441989 |
1
|
16
|
|
GO:0016810
|
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds
|
0.0507099 |
0.0507099 |
1
|
100
|
|
GO:0046112
|
nucleobase biosynthetic process
|
0.0595238 |
0.0595238 |
1
|
10
|
|
GO:0051726
|
regulation of cell cycle
|
0.0752212 |
0.0752212 |
1
|
187
|
|
GO:0055092
|
sterol homeostasis
|
0.0769231 |
0.0769231 |
1
|
1
|
|
GO:0034654
|
nucleobase, nucleoside, nucleotide and nucleic acid biosynthetic process
|
0.0870726 |
0.0870726 |
1
|
163
|
|
GO:0044271
|
cellular nitrogen compound biosynthetic process
|
0.0931953 |
0.0931953 |
1
|
252
|
|
GO:0044283
|
small molecule biosynthetic process
|
0.105199 |
0.105199 |
1
|
259
|
|
GO:0048522
|
positive regulation of cellular process
|
0.110028 |
0.110028 |
1
|
372
|
|
GO:0048518
|
positive regulation of biological process
|
0.111456 |
0.111456 |
1
|
410
|
|
GO:0055086
|
nucleobase, nucleoside and nucleotide metabolic process
|
0.112568 |
0.112568 |
1
|
249
|
|
GO:0046483
|
heterocycle metabolic process
|
0.147277 |
0.147277 |
1
|
292
|
|
GO:0055088
|
lipid homeostasis
|
0.164557 |
0.164557 |
1
|
13
|
|
GO:0043412
|
macromolecule modification
|
0.201784 |
0.201784 |
1
|
724
|
|
GO:0007049
|
cell cycle
|
0.217222 |
0.217222 |
1
|
756
|
|
GO:0044237
|
cellular metabolic process
|
0.241038 |
0.241038 |
2
|
3814
|
|
GO:0004157
|
dihydropyrimidinase activity
|
0.250000 |
0.250000 |
1
|
1
|
|
GO:0042592
|
homeostatic process
|
0.256516 |
0.256516 |
1
|
187
|
|
GO:0043167
|
ion binding
|
0.265556 |
0.265556 |
1
|
1498
|
|
GO:0006793
|
phosphorus metabolic process
|
0.270339 |
0.270339 |
1
|
556
|
|
GO:0044281
|
small molecule metabolic process
|
0.271588 |
0.271588 |
1
|
761
|
|
GO:0065008
|
regulation of biological quality
|
0.286107 |
0.286107 |
1
|
729
|
|
GO:0003824
|
catalytic activity
|
0.310936 |
0.310936 |
2
|
4106
|
|
GO:0006468
|
protein phosphorylation
|
0.335758 |
0.335758 |
1
|
277
|
|
GO:0048878
|
chemical homeostasis
|
0.422460 |
0.422460 |
1
|
79
|
|
GO:0016740
|
transferase activity
|
0.438824 |
0.438824 |
1
|
1030
|
|
GO:0044267
|
cellular protein metabolic process
|
0.440058 |
0.440058 |
1
|
1505
|
|
GO:0006464
|
protein modification process
|
0.443005 |
0.443005 |
1
|
684
|
|
GO:0019856
|
pyrimidine base biosynthetic process
|
0.461538 |
0.461538 |
1
|
6
|
|
GO:0034404
|
nucleobase, nucleoside and nucleotide biosynthetic process
|
0.473837 |
0.473837 |
1
|
163
|
|
GO:0016772
|
transferase activity, transferring phosphorus-containing groups
|
0.480583 |
0.480583 |
1
|
495
|
|
GO:0008152
|
metabolic process
|
0.483893 |
0.483893 |
2
|
5194
|
|
GO:0050789
|
regulation of biological process
|
0.509927 |
0.509927 |
1
|
2246
|
|
GO:0050794
|
regulation of cellular process
|
0.518911 |
0.518911 |
1
|
2034
|
|
GO:0065007
|
biological regulation
|
0.561089 |
0.561089 |
1
|
2548
|
|
GO:0006206
|
pyrimidine base metabolic process
|
0.562500 |
0.562500 |
1
|
9
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0009058
|
biosynthetic process
|
0.642905 |
0.642905 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.643595 |
0.643595 |
1
|
2093
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.658716 |
0.658716 |
1
|
1789
|
|
GO:0044238
|
primary metabolic process
|
0.672346 |
0.672346 |
2
|
4259
|
|
GO:0009987
|
cellular process
|
0.673638 |
0.673638 |
2
|
6560
|
|
GO:0004674
|
protein serine/threonine kinase activity
|
0.686207 |
0.686207 |
1
|
199
|
|
GO:0005623
|
cell
|
0.702620 |
0.702620 |
1
|
6195
|
|
GO:0044464
|
cell part
|
0.702620 |
0.702620 |
1
|
6195
|
|
GO:0019538
|
protein metabolic process
|
0.724907 |
0.724907 |
1
|
2053
|
|
GO:0004672
|
protein kinase activity
|
0.728643 |
0.728643 |
1
|
290
|
|
GO:0016787
|
hydrolase activity
|
0.729944 |
0.729944 |
1
|
1972
|
|
GO:0016773
|
phosphotransferase activity, alcohol group as acceptor
|
0.735354 |
0.735354 |
1
|
364
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.748407 |
0.748407 |
1
|
1959
|
|
GO:0016310
|
phosphorylation
|
0.758993 |
0.758993 |
1
|
422
|
|
GO:0005622
|
intracellular
|
0.764165 |
0.764165 |
1
|
4734
|
|
GO:0044249
|
cellular biosynthetic process
|
0.776153 |
0.776153 |
1
|
2034
|
|
GO:0008270
|
zinc ion binding
|
0.781440 |
0.781440 |
1
|
901
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0016301
|
kinase activity
|
0.795960 |
0.795960 |
1
|
394
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.869824 |
0.869824 |
1
|
2878
|
|
GO:0005488
|
binding
|
0.882277 |
0.882277 |
1
|
5641
|
|
GO:0043170
|
macromolecule metabolic process
|
0.904435 |
0.904435 |
1
|
3588
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0005575
|
cellular_component
|
0.970710 |
0.970710 |
1
|
8817
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0006207
|
'de novo' pyrimidine base biosynthetic process
|
1.00000 |
1.00000 |
1
|
6
|
|
GO:0006796
|
phosphate metabolic process
|
1.00000 |
1.00000 |
1
|
556
|
|
GO:0042632
|
cholesterol homeostasis
|
1.00000 |
1.00000 |
1
|
1
|