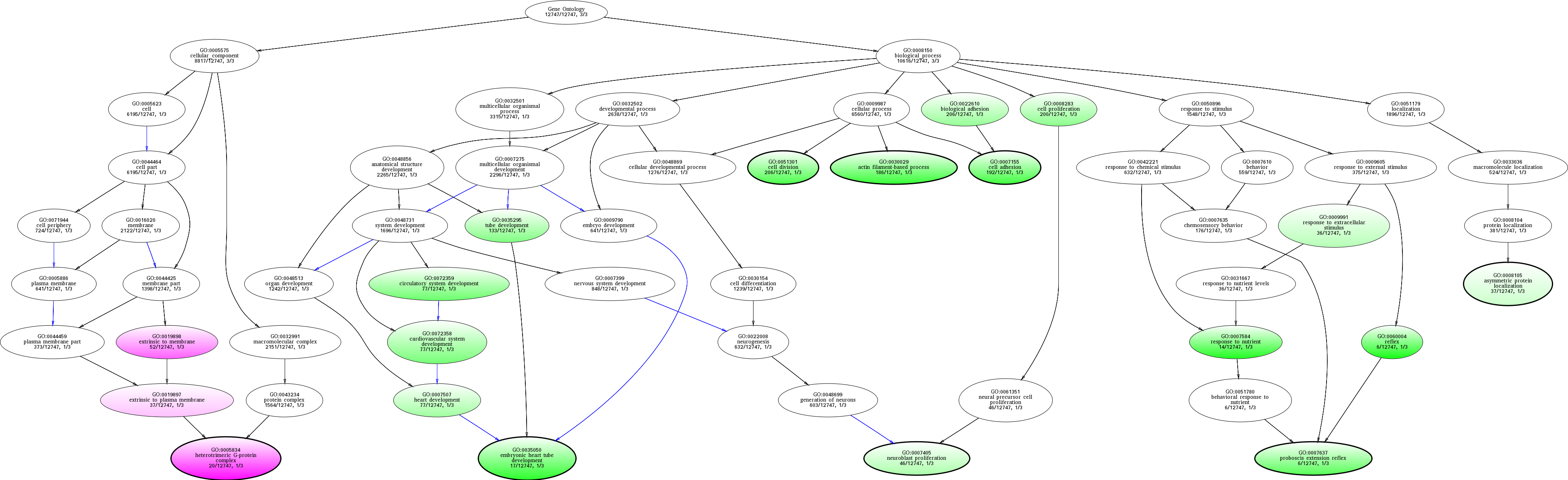
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0005834
|
heterotrimeric G-protein complex
|
0.0127389 |
0.0127389 |
1
|
20
|
|
GO:0060004
|
reflex
|
0.0160000 |
0.0160000 |
1
|
6
|
|
GO:0007584
|
response to nutrient
|
0.0217054 |
0.0217054 |
1
|
14
|
|
GO:0035050
|
embryonic heart tube development
|
0.0236111 |
0.0236111 |
1
|
17
|
|
GO:0030029
|
actin filament-based process
|
0.0283537 |
0.0283537 |
1
|
186
|
|
GO:0007155
|
cell adhesion
|
0.0292104 |
0.0292104 |
1
|
192
|
|
GO:0051301
|
cell division
|
0.0314024 |
0.0314024 |
1
|
206
|
|
GO:0007637
|
proboscis extension reflex
|
0.0340909 |
0.0340909 |
1
|
6
|
|
GO:0019898
|
extrinsic to membrane
|
0.0371960 |
0.0371960 |
1
|
52
|
|
GO:0072359
|
circulatory system development
|
0.0454009 |
0.0454009 |
1
|
77
|
|
GO:0072358
|
cardiovascular system development
|
0.0454009 |
0.0454009 |
1
|
77
|
|
GO:0035295
|
tube development
|
0.0515704 |
0.0515704 |
1
|
133
|
|
GO:0008283
|
cell proliferation
|
0.0554655 |
0.0554655 |
1
|
200
|
|
GO:0022610
|
biological adhesion
|
0.0570970 |
0.0570970 |
1
|
206
|
|
GO:0007507
|
heart development
|
0.0619968 |
0.0619968 |
1
|
77
|
|
GO:0007405
|
neuroblast proliferation
|
0.0762852 |
0.0762852 |
1
|
46
|
|
GO:0009991
|
response to extracellular stimulus
|
0.0960000 |
0.0960000 |
1
|
36
|
|
GO:0019897
|
extrinsic to plasma membrane
|
0.0963542 |
0.0963542 |
1
|
37
|
|
GO:0008105
|
asymmetric protein localization
|
0.0971129 |
0.0971129 |
1
|
37
|
|
GO:0071944
|
cell periphery
|
0.116868 |
0.116868 |
1
|
724
|
|
GO:0055059
|
asymmetric neuroblast division
|
0.117647 |
0.117647 |
1
|
30
|
|
GO:0060089
|
molecular transducer activity
|
0.144056 |
0.144056 |
1
|
559
|
|
GO:0008360
|
regulation of cell shape
|
0.153635 |
0.153635 |
1
|
112
|
|
GO:0048869
|
cellular developmental process
|
0.176780 |
0.176780 |
1
|
1276
|
|
GO:0007635
|
chemosensory behavior
|
0.186441 |
0.186441 |
1
|
176
|
|
GO:0045165
|
cell fate commitment
|
0.199843 |
0.199843 |
1
|
255
|
|
GO:0044425
|
membrane part
|
0.225666 |
0.225666 |
1
|
1398
|
|
GO:0061351
|
neural precursor cell proliferation
|
0.230000 |
0.230000 |
1
|
46
|
|
GO:0044459
|
plasma membrane part
|
0.236675 |
0.236675 |
1
|
373
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.237386 |
0.237386 |
1
|
1562
|
|
GO:0003924
|
GTPase activity
|
0.241796 |
0.241796 |
1
|
140
|
|
GO:0009605
|
response to external stimulus
|
0.242248 |
0.242248 |
1
|
375
|
|
GO:0009790
|
embryo development
|
0.242987 |
0.242987 |
1
|
641
|
|
GO:0045176
|
apical protein localization
|
0.243243 |
0.243243 |
1
|
9
|
|
GO:0033036
|
macromolecule localization
|
0.276371 |
0.276371 |
1
|
524
|
|
GO:0065008
|
regulation of biological quality
|
0.286107 |
0.286107 |
1
|
729
|
|
GO:0005886
|
plasma membrane
|
0.292027 |
0.292027 |
1
|
641
|
|
GO:0016817
|
hydrolase activity, acting on acid anhydrides
|
0.301217 |
0.301217 |
1
|
594
|
|
GO:0007186
|
G-protein coupled receptor protein signaling pathway
|
0.318910 |
0.318910 |
1
|
199
|
|
GO:0005575
|
cellular_component
|
0.330897 |
0.330897 |
3
|
8817
|
|
GO:0023052
|
signaling
|
0.338079 |
0.338079 |
1
|
1364
|
|
GO:0030036
|
actin cytoskeleton organization
|
0.339518 |
0.339518 |
1
|
183
|
|
GO:0017145
|
stem cell division
|
0.339806 |
0.339806 |
1
|
70
|
|
GO:0016020
|
membrane
|
0.342534 |
0.342534 |
1
|
2122
|
|
GO:0007610
|
behavior
|
0.361111 |
0.361111 |
1
|
559
|
|
GO:0050896
|
response to stimulus
|
0.376795 |
0.376795 |
1
|
1548
|
|
GO:0042221
|
response to chemical stimulus
|
0.408269 |
0.408269 |
1
|
632
|
|
GO:0051780
|
behavioral response to nutrient
|
0.428571 |
0.428571 |
1
|
6
|
|
GO:0051179
|
localization
|
0.445834 |
0.445834 |
1
|
1896
|
|
GO:0022008
|
neurogenesis
|
0.452398 |
0.452398 |
1
|
632
|
|
GO:0007010
|
cytoskeleton organization
|
0.453469 |
0.453469 |
1
|
536
|
|
GO:0016787
|
hydrolase activity
|
0.480273 |
0.480273 |
1
|
1972
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.485282 |
0.485282 |
1
|
2108
|
|
GO:0007399
|
nervous system development
|
0.500000 |
0.500000 |
1
|
848
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0048103
|
somatic stem cell division
|
0.557143 |
0.557143 |
1
|
39
|
|
GO:0065007
|
biological regulation
|
0.561089 |
0.561089 |
1
|
2548
|
|
GO:0032991
|
macromolecular complex
|
0.567899 |
0.567899 |
1
|
2151
|
|
GO:0055057
|
neuroblast division
|
0.574074 |
0.574074 |
1
|
31
|
|
GO:0032502
|
developmental process
|
0.575616 |
0.575616 |
1
|
2638
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0007015
|
actin filament organization
|
0.606557 |
0.606557 |
1
|
111
|
|
GO:0007166
|
cell surface receptor linked signaling pathway
|
0.652720 |
0.652720 |
1
|
624
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0032501
|
multicellular organismal process
|
0.674757 |
0.674757 |
1
|
3315
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.724105 |
0.724105 |
1
|
1517
|
|
GO:0008104
|
protein localization
|
0.727099 |
0.727099 |
1
|
381
|
|
GO:0043234
|
protein complex
|
0.727104 |
0.727104 |
1
|
1564
|
|
GO:0003824
|
catalytic activity
|
0.751316 |
0.751316 |
1
|
4106
|
|
GO:0006996
|
organelle organization
|
0.779169 |
0.779169 |
1
|
1182
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0009987
|
cellular process
|
0.944254 |
0.944254 |
1
|
6560
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0005623
|
cell
|
0.973722 |
0.973722 |
1
|
6195
|
|
GO:0044464
|
cell part
|
0.973722 |
0.973722 |
1
|
6195
|
|
GO:0017111
|
nucleoside-triphosphatase activity
|
0.983022 |
0.983022 |
1
|
579
|
|
GO:0016462
|
pyrophosphatase activity
|
0.991582 |
0.991582 |
1
|
589
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0016818
|
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
|
1.00000 |
1.00000 |
1
|
594
|
|
GO:0004871
|
signal transducer activity
|
1.00000 |
1.00000 |
1
|
559
|
|
GO:0031667
|
response to nutrient levels
|
1.00000 |
1.00000 |
1
|
36
|