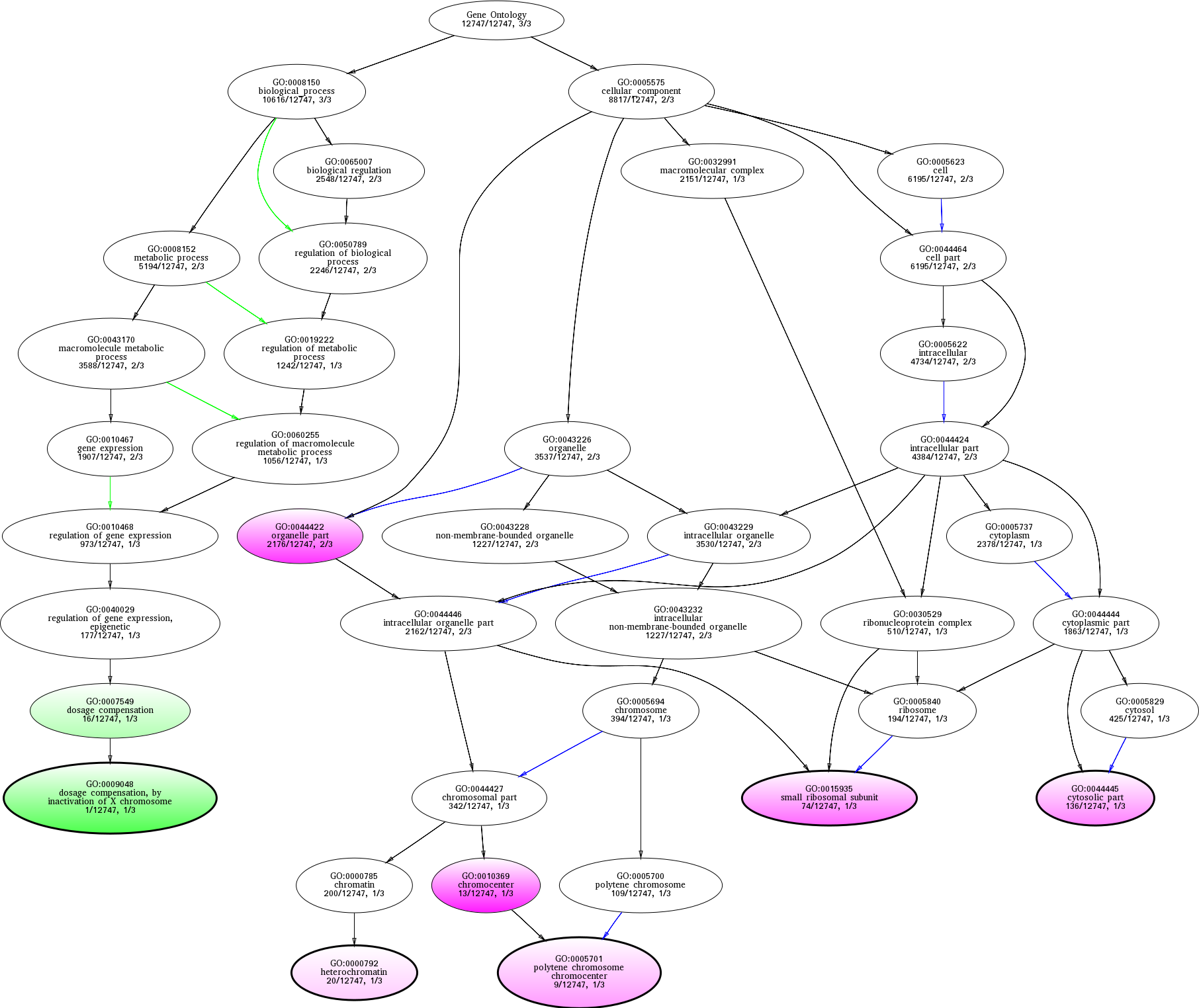
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0010369
|
chromocenter
|
0.0380117 |
0.0380117 |
1
|
13
|
|
GO:0044422
|
organelle part
|
0.0608872 |
0.0608872 |
2
|
2176
|
|
GO:0009048
|
dosage compensation, by inactivation of X chromosome
|
0.0625000 |
0.0625000 |
1
|
1
|
|
GO:0015935
|
small ribosomal subunit
|
0.0655991 |
0.0655991 |
1
|
74
|
|
GO:0044445
|
cytosolic part
|
0.0730005 |
0.0730005 |
1
|
136
|
|
GO:0005701
|
polytene chromosome chromocenter
|
0.0810811 |
0.0810811 |
1
|
9
|
|
GO:0007549
|
dosage compensation
|
0.0903955 |
0.0903955 |
1
|
16
|
|
GO:0000792
|
heterochromatin
|
0.100000 |
0.100000 |
1
|
20
|
|
GO:0050789
|
regulation of biological process
|
0.115315 |
0.115315 |
2
|
2246
|
|
GO:0005198
|
structural molecule activity
|
0.119376 |
0.119376 |
1
|
459
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.120278 |
0.120278 |
2
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.120756 |
0.120756 |
2
|
1227
|
|
GO:0005840
|
ribosome
|
0.131663 |
0.131663 |
1
|
194
|
|
GO:0005811
|
lipid particle
|
0.134192 |
0.134192 |
1
|
250
|
|
GO:0065007
|
biological regulation
|
0.145142 |
0.145142 |
2
|
2548
|
|
GO:0043226
|
organelle
|
0.160900 |
0.160900 |
2
|
3537
|
|
GO:0000775
|
chromosome, centromeric region
|
0.163743 |
0.163743 |
1
|
56
|
|
GO:0040029
|
regulation of gene expression, epigenetic
|
0.181912 |
0.181912 |
1
|
177
|
|
GO:0019001
|
guanyl nucleotide binding
|
0.182875 |
0.182875 |
1
|
173
|
|
GO:0032561
|
guanyl ribonucleotide binding
|
0.194098 |
0.194098 |
1
|
171
|
|
GO:0023046
|
signaling process
|
0.199779 |
0.199779 |
1
|
760
|
|
GO:0030529
|
ribonucleoprotein complex
|
0.210589 |
0.210589 |
1
|
510
|
|
GO:0005829
|
cytosol
|
0.228127 |
0.228127 |
1
|
425
|
|
GO:0003924
|
GTPase activity
|
0.241796 |
0.241796 |
1
|
140
|
|
GO:0044446
|
intracellular organelle part
|
0.243036 |
0.243036 |
2
|
2162
|
|
GO:0007165
|
signal transduction
|
0.254057 |
0.254057 |
1
|
548
|
|
GO:0005700
|
polytene chromosome
|
0.276650 |
0.276650 |
1
|
109
|
|
GO:0010467
|
gene expression
|
0.282416 |
0.282416 |
2
|
1907
|
|
GO:0044427
|
chromosomal part
|
0.288122 |
0.288122 |
1
|
342
|
|
GO:0016817
|
hydrolase activity, acting on acid anhydrides
|
0.301217 |
0.301217 |
1
|
594
|
|
GO:0023052
|
signaling
|
0.338079 |
0.338079 |
1
|
1364
|
|
GO:0022627
|
cytosolic small ribosomal subunit
|
0.338462 |
0.338462 |
1
|
44
|
|
GO:0000166
|
nucleotide binding
|
0.374077 |
0.374077 |
1
|
1178
|
|
GO:0006412
|
translation
|
0.382593 |
0.382593 |
1
|
600
|
|
GO:0003735
|
structural constituent of ribosome
|
0.398693 |
0.398693 |
1
|
183
|
|
GO:0032991
|
macromolecular complex
|
0.428425 |
0.428425 |
1
|
2151
|
|
GO:0023034
|
intracellular signaling pathway
|
0.430962 |
0.430962 |
1
|
412
|
|
GO:0022626
|
cytosolic ribosome
|
0.434783 |
0.434783 |
1
|
100
|
|
GO:0044267
|
cellular protein metabolic process
|
0.440058 |
0.440058 |
1
|
1505
|
|
GO:0003677
|
DNA binding
|
0.444205 |
0.444205 |
1
|
824
|
|
GO:0043167
|
ion binding
|
0.460626 |
0.460626 |
1
|
1498
|
|
GO:0043170
|
macromolecule metabolic process
|
0.477159 |
0.477159 |
2
|
3588
|
|
GO:0016787
|
hydrolase activity
|
0.480273 |
0.480273 |
1
|
1972
|
|
GO:0008152
|
metabolic process
|
0.483893 |
0.483893 |
2
|
5194
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.486014 |
0.486014 |
1
|
1056
|
|
GO:0005623
|
cell
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.496927 |
0.496927 |
1
|
1617
|
|
GO:0035556
|
intracellular signal transduction
|
0.499208 |
0.499208 |
1
|
315
|
|
GO:0044424
|
intracellular part
|
0.500760 |
0.500760 |
2
|
4384
|
|
GO:0019222
|
regulation of metabolic process
|
0.507779 |
0.507779 |
1
|
1242
|
|
GO:0005488
|
binding
|
0.514777 |
0.514777 |
2
|
5641
|
|
GO:0005515
|
protein binding
|
0.518366 |
0.518366 |
1
|
1726
|
|
GO:0044249
|
cellular biosynthetic process
|
0.526807 |
0.526807 |
1
|
2034
|
|
GO:0005694
|
chromosome
|
0.539284 |
0.539284 |
1
|
394
|
|
GO:0003676
|
nucleic acid binding
|
0.549587 |
0.549587 |
1
|
1855
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0005622
|
intracellular
|
0.583919 |
0.583919 |
2
|
4734
|
|
GO:0000785
|
chromatin
|
0.584795 |
0.584795 |
1
|
200
|
|
GO:0007264
|
small GTPase mediated signal transduction
|
0.587302 |
0.587302 |
1
|
185
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0009058
|
biosynthetic process
|
0.642905 |
0.642905 |
1
|
2090
|
|
GO:0043229
|
intracellular organelle
|
0.646541 |
0.646541 |
2
|
3530
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.646807 |
0.646807 |
1
|
1623
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0050794
|
regulation of cellular process
|
0.666348 |
0.666348 |
1
|
2034
|
|
GO:0044444
|
cytoplasmic part
|
0.669378 |
0.669378 |
1
|
1863
|
|
GO:0009987
|
cellular process
|
0.673638 |
0.673638 |
2
|
6560
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0019538
|
protein metabolic process
|
0.724907 |
0.724907 |
1
|
2053
|
|
GO:0010468
|
regulation of gene expression
|
0.739954 |
0.739954 |
1
|
973
|
|
GO:0032553
|
ribonucleotide binding
|
0.746180 |
0.746180 |
1
|
879
|
|
GO:0003824
|
catalytic activity
|
0.751316 |
0.751316 |
1
|
4106
|
|
GO:0005575
|
cellular_component
|
0.773470 |
0.773470 |
2
|
8817
|
|
GO:0008270
|
zinc ion binding
|
0.781440 |
0.781440 |
1
|
901
|
|
GO:0005737
|
cytoplasm
|
0.790684 |
0.790684 |
1
|
2378
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0017076
|
purine nucleotide binding
|
0.803056 |
0.803056 |
1
|
946
|
|
GO:0044237
|
cellular metabolic process
|
0.868168 |
0.868168 |
1
|
3814
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.869824 |
0.869824 |
1
|
2878
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.929175 |
0.929175 |
1
|
879
|
|
GO:0043227
|
membrane-bounded organelle
|
0.963300 |
0.963300 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.963413 |
0.963413 |
1
|
2856
|
|
GO:0044238
|
primary metabolic process
|
0.967623 |
0.967623 |
1
|
4259
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0017111
|
nucleoside-triphosphatase activity
|
0.983022 |
0.983022 |
1
|
579
|
|
GO:0016462
|
pyrophosphatase activity
|
0.991582 |
0.991582 |
1
|
589
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0016818
|
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
|
1.00000 |
1.00000 |
1
|
594
|
|
GO:0005525
|
GTP binding
|
1.00000 |
1.00000 |
1
|
171
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|