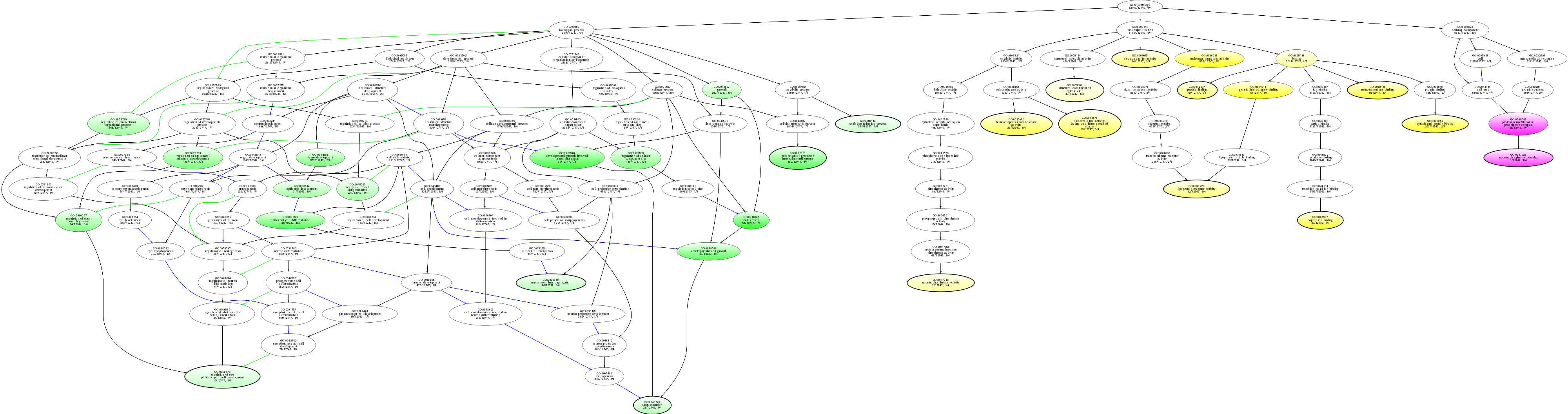
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0071814
|
protein-lipid complex binding
|
0.00918884 |
0.00918884 |
1
|
13
|
|
GO:0060089
|
molecular transducer activity
|
0.0142830 |
0.0142830 |
2
|
559
|
|
GO:0008092
|
cytoskeletal protein binding
|
0.0161822 |
0.0161822 |
2
|
220
|
|
GO:0008287
|
protein serine/threonine phosphatase complex
|
0.0179095 |
0.0179095 |
1
|
28
|
|
GO:0005507
|
copper ion binding
|
0.0199480 |
0.0199480 |
1
|
23
|
|
GO:0016049
|
cell growth
|
0.0274662 |
0.0274662 |
1
|
91
|
|
GO:0042165
|
neurotransmitter binding
|
0.0343045 |
0.0343045 |
1
|
49
|
|
GO:0060560
|
developmental growth involved in morphogenesis
|
0.0355105 |
0.0355105 |
1
|
56
|
|
GO:0017023
|
myosin phosphatase complex
|
0.0357143 |
0.0357143 |
1
|
1
|
|
GO:0016675
|
oxidoreductase activity, acting on a heme group of donors
|
0.0360502 |
0.0360502 |
1
|
23
|
|
GO:0015002
|
heme-copper terminal oxidase activity
|
0.0360502 |
0.0360502 |
1
|
23
|
|
GO:0009913
|
epidermal cell differentiation
|
0.0369775 |
0.0369775 |
1
|
46
|
|
GO:0048588
|
developmental cell growth
|
0.0389493 |
0.0389493 |
1
|
43
|
|
GO:0006091
|
generation of precursor metabolites and energy
|
0.0398532 |
0.0398532 |
1
|
152
|
|
GO:0042277
|
peptide binding
|
0.0439416 |
0.0439416 |
1
|
63
|
|
GO:0009055
|
electron carrier activity
|
0.0579938 |
0.0579938 |
1
|
164
|
|
GO:0030228
|
lipoprotein receptor activity
|
0.0597622 |
0.0597622 |
1
|
12
|
|
GO:0009888
|
tissue development
|
0.0603928 |
0.0603928 |
2
|
557
|
|
GO:0022603
|
regulation of anatomical structure morphogenesis
|
0.0618812 |
0.0618812 |
1
|
100
|
|
GO:0032535
|
regulation of cellular component size
|
0.0631681 |
0.0631681 |
1
|
130
|
|
GO:2000027
|
regulation of organ morphogenesis
|
0.0659341 |
0.0659341 |
1
|
54
|
|
GO:0005488
|
binding
|
0.0675382 |
0.0675382 |
4
|
5641
|
|
GO:0017018
|
myosin phosphatase activity
|
0.0697674 |
0.0697674 |
1
|
3
|
|
GO:0051239
|
regulation of multicellular organismal process
|
0.0844424 |
0.0844424 |
1
|
368
|
|
GO:0008544
|
epidermis development
|
0.0858137 |
0.0858137 |
1
|
61
|
|
GO:0045595
|
regulation of cell differentiation
|
0.0877660 |
0.0877660 |
1
|
231
|
|
GO:0040007
|
growth
|
0.0898814 |
0.0898814 |
1
|
247
|
|
GO:0035316
|
non-sensory hair organization
|
0.0907216 |
0.0907216 |
1
|
44
|
|
GO:0048675
|
axon extension
|
0.0924092 |
0.0924092 |
1
|
28
|
|
GO:0005200
|
structural constituent of cytoskeleton
|
0.0936819 |
0.0936819 |
1
|
43
|
|
GO:0042478
|
regulation of eye photoreceptor cell development
|
0.0973451 |
0.0973451 |
1
|
11
|
|
GO:0055114
|
oxidation-reduction process
|
0.0993454 |
0.0993454 |
1
|
516
|
|
GO:2000026
|
regulation of multicellular organismal development
|
0.109792 |
0.109792 |
1
|
259
|
|
GO:0048589
|
developmental growth
|
0.112049 |
0.112049 |
1
|
154
|
|
GO:0046532
|
regulation of photoreceptor cell differentiation
|
0.118812 |
0.118812 |
1
|
24
|
|
GO:0045314
|
regulation of compound eye photoreceptor development
|
0.123457 |
0.123457 |
1
|
10
|
|
GO:0050767
|
regulation of neurogenesis
|
0.131004 |
0.131004 |
1
|
90
|
|
GO:0045664
|
regulation of neuron differentiation
|
0.131907 |
0.131907 |
1
|
74
|
|
GO:0051960
|
regulation of nervous system development
|
0.132196 |
0.132196 |
1
|
124
|
|
GO:0004983
|
neuropeptide Y receptor activity
|
0.133333 |
0.133333 |
1
|
6
|
|
GO:0048598
|
embryonic morphogenesis
|
0.136979 |
0.136979 |
1
|
243
|
|
GO:0030030
|
cell projection organization
|
0.142264 |
0.142264 |
1
|
485
|
|
GO:0022900
|
electron transport chain
|
0.143911 |
0.143911 |
1
|
78
|
|
GO:0005509
|
calcium ion binding
|
0.144237 |
0.144237 |
1
|
209
|
|
GO:0042461
|
photoreceptor cell development
|
0.147601 |
0.147601 |
1
|
80
|
|
GO:0005198
|
structural molecule activity
|
0.155920 |
0.155920 |
1
|
459
|
|
GO:0060284
|
regulation of cell development
|
0.159091 |
0.159091 |
1
|
168
|
|
GO:0050793
|
regulation of developmental process
|
0.161873 |
0.161873 |
1
|
321
|
|
GO:0035317
|
imaginal disc-derived wing hair organization
|
0.168627 |
0.168627 |
1
|
43
|
|
GO:0035107
|
appendage morphogenesis
|
0.184365 |
0.184365 |
1
|
283
|
|
GO:0001653
|
peptide receptor activity
|
0.192469 |
0.192469 |
1
|
48
|
|
GO:0032989
|
cellular component morphogenesis
|
0.193108 |
0.193108 |
1
|
566
|
|
GO:0030594
|
neurotransmitter receptor activity
|
0.198011 |
0.198011 |
1
|
48
|
|
GO:0017022
|
myosin binding
|
0.198589 |
0.198589 |
1
|
23
|
|
GO:0008528
|
peptide receptor activity, G-protein coupled
|
0.199170 |
0.199170 |
1
|
48
|
|
GO:0048736
|
appendage development
|
0.209532 |
0.209532 |
1
|
286
|
|
GO:0048676
|
axon extension involved in development
|
0.214286 |
0.214286 |
1
|
6
|
|
GO:0044425
|
membrane part
|
0.221352 |
0.221352 |
2
|
1398
|
|
GO:0005575
|
cellular_component
|
0.228855 |
0.228855 |
4
|
8817
|
|
GO:0004129
|
cytochrome-c oxidase activity
|
0.230000 |
0.230000 |
1
|
23
|
|
GO:0044430
|
cytoskeletal part
|
0.231674 |
0.231674 |
1
|
512
|
|
GO:0090066
|
regulation of anatomical structure size
|
0.231824 |
0.231824 |
1
|
169
|
|
GO:0005623
|
cell
|
0.243645 |
0.243645 |
4
|
6195
|
|
GO:0044464
|
cell part
|
0.243645 |
0.243645 |
4
|
6195
|
|
GO:0016788
|
hydrolase activity, acting on ester bonds
|
0.251521 |
0.251521 |
1
|
496
|
|
GO:0032991
|
macromolecular complex
|
0.251544 |
0.251544 |
2
|
2151
|
|
GO:0032502
|
developmental process
|
0.259158 |
0.259158 |
2
|
2638
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.259446 |
0.259446 |
1
|
412
|
|
GO:0007552
|
metamorphosis
|
0.273794 |
0.273794 |
1
|
420
|
|
GO:0005215
|
transporter activity
|
0.274270 |
0.274270 |
1
|
852
|
|
GO:0048569
|
post-embryonic organ development
|
0.276167 |
0.276167 |
1
|
343
|
|
GO:0048609
|
multicellular organismal reproductive process
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0032504
|
multicellular organism reproduction
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0065008
|
regulation of biological quality
|
0.286107 |
0.286107 |
1
|
729
|
|
GO:0016491
|
oxidoreductase activity
|
0.286653 |
0.286653 |
1
|
638
|
|
GO:0046530
|
photoreceptor cell differentiation
|
0.295620 |
0.295620 |
1
|
162
|
|
GO:0007498
|
mesoderm development
|
0.312278 |
0.312278 |
1
|
95
|
|
GO:0007186
|
G-protein coupled receptor protein signaling pathway
|
0.318910 |
0.318910 |
1
|
199
|
|
GO:0000003
|
reproduction
|
0.332995 |
0.332995 |
1
|
1022
|
|
GO:0022414
|
reproductive process
|
0.332995 |
0.332995 |
1
|
1022
|
|
GO:0009791
|
post-embryonic development
|
0.343209 |
0.343209 |
1
|
500
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.346904 |
0.346904 |
1
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.347592 |
0.347592 |
1
|
1227
|
|
GO:0016331
|
morphogenesis of embryonic epithelium
|
0.347709 |
0.347709 |
1
|
129
|
|
GO:0048729
|
tissue morphogenesis
|
0.350704 |
0.350704 |
1
|
312
|
|
GO:0001751
|
compound eye photoreceptor cell differentiation
|
0.354497 |
0.354497 |
1
|
134
|
|
GO:0005515
|
protein binding
|
0.358840 |
0.358840 |
2
|
1726
|
|
GO:0001754
|
eye photoreceptor cell differentiation
|
0.361386 |
0.361386 |
1
|
146
|
|
GO:0009887
|
organ morphogenesis
|
0.374384 |
0.374384 |
1
|
684
|
|
GO:0009792
|
embryo development ending in birth or egg hatching
|
0.374415 |
0.374415 |
1
|
240
|
|
GO:0007444
|
imaginal disc development
|
0.376006 |
0.376006 |
1
|
467
|
|
GO:0000904
|
cell morphogenesis involved in differentiation
|
0.376731 |
0.376731 |
1
|
408
|
|
GO:0007391
|
dorsal closure
|
0.392996 |
0.392996 |
1
|
101
|
|
GO:0005875
|
microtubule associated complex
|
0.399123 |
0.399123 |
1
|
363
|
|
GO:0048563
|
post-embryonic organ morphogenesis
|
0.415167 |
0.415167 |
1
|
323
|
|
GO:0023052
|
signaling
|
0.423150 |
0.423150 |
1
|
1364
|
|
GO:0016020
|
membrane
|
0.423766 |
0.423766 |
2
|
2122
|
|
GO:0009790
|
embryo development
|
0.427001 |
0.427001 |
1
|
641
|
|
GO:0048666
|
neuron development
|
0.433702 |
0.433702 |
1
|
471
|
|
GO:0004722
|
protein serine/threonine phosphatase activity
|
0.434343 |
0.434343 |
1
|
43
|
|
GO:0007423
|
sensory organ development
|
0.438003 |
0.438003 |
1
|
544
|
|
GO:0042578
|
phosphoric ester hydrolase activity
|
0.441532 |
0.441532 |
1
|
219
|
|
GO:0048869
|
cellular developmental process
|
0.442161 |
0.442161 |
1
|
1276
|
|
GO:0030182
|
neuron differentiation
|
0.442292 |
0.442292 |
1
|
548
|
|
GO:0022008
|
neurogenesis
|
0.452398 |
0.452398 |
1
|
632
|
|
GO:0005856
|
cytoskeleton
|
0.457213 |
0.457213 |
1
|
561
|
|
GO:0042462
|
eye photoreceptor cell development
|
0.458065 |
0.458065 |
1
|
71
|
|
GO:0003824
|
catalytic activity
|
0.474372 |
0.474372 |
2
|
4106
|
|
GO:0008150
|
biological_process
|
0.481028 |
0.481028 |
4
|
10616
|
|
GO:0044446
|
intracellular organelle part
|
0.493044 |
0.493044 |
1
|
2162
|
|
GO:0007399
|
nervous system development
|
0.500000 |
0.500000 |
1
|
848
|
|
GO:0022890
|
inorganic cation transmembrane transporter activity
|
0.502439 |
0.502439 |
1
|
206
|
|
GO:0042051
|
compound eye photoreceptor development
|
0.507353 |
0.507353 |
1
|
69
|
|
GO:0050794
|
regulation of cellular process
|
0.518911 |
0.518911 |
1
|
2034
|
|
GO:0004721
|
phosphoprotein phosphatase activity
|
0.523810 |
0.523810 |
1
|
99
|
|
GO:0031224
|
intrinsic to membrane
|
0.525949 |
0.525949 |
2
|
1014
|
|
GO:0043234
|
protein complex
|
0.528587 |
0.528587 |
2
|
1564
|
|
GO:0048592
|
eye morphogenesis
|
0.542069 |
0.542069 |
1
|
393
|
|
GO:0003674
|
molecular_function
|
0.567526 |
0.567526 |
4
|
11064
|
|
GO:0015078
|
hydrogen ion transmembrane transporter activity
|
0.574713 |
0.574713 |
1
|
100
|
|
GO:0007560
|
imaginal disc morphogenesis
|
0.575758 |
0.575758 |
1
|
323
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.587518 |
0.587518 |
1
|
2108
|
|
GO:0050789
|
regulation of biological process
|
0.613640 |
0.613640 |
1
|
2246
|
|
GO:0035120
|
post-embryonic appendage morphogenesis
|
0.638095 |
0.638095 |
1
|
268
|
|
GO:0007166
|
cell surface receptor linked signaling pathway
|
0.652720 |
0.652720 |
1
|
624
|
|
GO:0007472
|
wing disc morphogenesis
|
0.665796 |
0.665796 |
1
|
255
|
|
GO:0065007
|
biological regulation
|
0.666464 |
0.666464 |
1
|
2548
|
|
GO:0004872
|
receptor activity
|
0.673957 |
0.673957 |
2
|
459
|
|
GO:0044422
|
organelle part
|
0.678224 |
0.678224 |
1
|
2176
|
|
GO:0007409
|
axonogenesis
|
0.679389 |
0.679389 |
1
|
267
|
|
GO:0035220
|
wing disc development
|
0.680942 |
0.680942 |
1
|
318
|
|
GO:0031175
|
neuron projection development
|
0.696270 |
0.696270 |
1
|
392
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0048468
|
cell development
|
0.705715 |
0.705715 |
1
|
1042
|
|
GO:0043167
|
ion binding
|
0.709151 |
0.709151 |
1
|
1498
|
|
GO:0042923
|
neuropeptide binding
|
0.714286 |
0.714286 |
1
|
45
|
|
GO:0007292
|
female gamete generation
|
0.727380 |
0.727380 |
1
|
619
|
|
GO:0016787
|
hydrolase activity
|
0.729944 |
0.729944 |
1
|
1972
|
|
GO:0048856
|
anatomical structure development
|
0.737157 |
0.737157 |
2
|
2265
|
|
GO:0004888
|
transmembrane receptor activity
|
0.740313 |
0.740313 |
2
|
395
|
|
GO:0044237
|
cellular metabolic process
|
0.740940 |
0.740940 |
1
|
3814
|
|
GO:0032990
|
cell part morphogenesis
|
0.745583 |
0.745583 |
1
|
422
|
|
GO:0008361
|
regulation of cell size
|
0.769231 |
0.769231 |
1
|
100
|
|
GO:0030169
|
low-density lipoprotein binding
|
0.769231 |
0.769231 |
1
|
10
|
|
GO:0032501
|
multicellular organismal process
|
0.776347 |
0.776347 |
1
|
3315
|
|
GO:0060429
|
epithelium development
|
0.779194 |
0.779194 |
1
|
295
|
|
GO:0022891
|
substrate-specific transmembrane transporter activity
|
0.788146 |
0.788146 |
1
|
625
|
|
GO:0003779
|
actin binding
|
0.794521 |
0.794521 |
1
|
120
|
|
GO:0048667
|
cell morphogenesis involved in neuron differentiation
|
0.795501 |
0.795501 |
1
|
389
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0048513
|
organ development
|
0.796117 |
0.796117 |
1
|
1242
|
|
GO:0015630
|
microtubule cytoskeleton
|
0.798574 |
0.798574 |
1
|
448
|
|
GO:0043229
|
intracellular organelle
|
0.804100 |
0.804100 |
1
|
3530
|
|
GO:0048858
|
cell projection morphogenesis
|
0.806883 |
0.806883 |
1
|
422
|
|
GO:0008324
|
cation transmembrane transporter activity
|
0.808679 |
0.808679 |
1
|
410
|
|
GO:0015075
|
ion transmembrane transporter activity
|
0.811200 |
0.811200 |
1
|
507
|
|
GO:0001745
|
compound eye morphogenesis
|
0.811947 |
0.811947 |
1
|
367
|
|
GO:0022892
|
substrate-specific transporter activity
|
0.815728 |
0.815728 |
1
|
695
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.824951 |
0.824951 |
1
|
1534
|
|
GO:0000902
|
cell morphogenesis
|
0.825088 |
0.825088 |
1
|
467
|
|
GO:0005041
|
low-density lipoprotein receptor activity
|
0.833333 |
0.833333 |
1
|
10
|
|
GO:0009987
|
cellular process
|
0.840883 |
0.840883 |
2
|
6560
|
|
GO:0001654
|
eye development
|
0.841912 |
0.841912 |
1
|
458
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.842767 |
0.842767 |
1
|
402
|
|
GO:0015077
|
monovalent inorganic cation transmembrane transporter activity
|
0.844660 |
0.844660 |
1
|
174
|
|
GO:0004930
|
G-protein coupled receptor activity
|
0.848602 |
0.848602 |
1
|
241
|
|
GO:0022857
|
transmembrane transporter activity
|
0.852113 |
0.852113 |
1
|
726
|
|
GO:0019953
|
sexual reproduction
|
0.859100 |
0.859100 |
1
|
878
|
|
GO:0016791
|
phosphatase activity
|
0.863014 |
0.863014 |
1
|
189
|
|
GO:0043226
|
organelle
|
0.871455 |
0.871455 |
1
|
3537
|
|
GO:0008188
|
neuropeptide receptor activity
|
0.882353 |
0.882353 |
1
|
45
|
|
GO:0048731
|
system development
|
0.882863 |
0.882863 |
1
|
1696
|
|
GO:0007276
|
gamete generation
|
0.883697 |
0.883697 |
1
|
851
|
|
GO:0007275
|
multicellular organismal development
|
0.887792 |
0.887792 |
1
|
2296
|
|
GO:0007476
|
imaginal disc-derived wing morphogenesis
|
0.903915 |
0.903915 |
1
|
254
|
|
GO:0002009
|
morphogenesis of an epithelium
|
0.908805 |
0.908805 |
1
|
289
|
|
GO:0048812
|
neuron projection morphogenesis
|
0.910798 |
0.910798 |
1
|
388
|
|
GO:0008152
|
metabolic process
|
0.931992 |
0.931992 |
1
|
5194
|
|
GO:0048749
|
compound eye development
|
0.932314 |
0.932314 |
1
|
427
|
|
GO:0002165
|
instar larval or pupal development
|
0.954000 |
0.954000 |
1
|
477
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0035315
|
hair cell differentiation
|
0.956522 |
0.956522 |
1
|
44
|
|
GO:0001700
|
embryonic development via the syncytial blastoderm
|
0.958333 |
0.958333 |
1
|
230
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0016021
|
integral to membrane
|
0.968675 |
0.968675 |
2
|
998
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0035114
|
imaginal disc-derived appendage morphogenesis
|
0.975524 |
0.975524 |
1
|
279
|
|
GO:0048737
|
imaginal disc-derived appendage development
|
0.986014 |
0.986014 |
1
|
282
|
|
GO:0048477
|
oogenesis
|
0.987076 |
0.987076 |
1
|
611
|
|
GO:0044424
|
intracellular part
|
0.992714 |
0.992714 |
1
|
4384
|
|
GO:0005622
|
intracellular
|
0.996916 |
0.996916 |
1
|
4734
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
4
|
12747
|
|
GO:0016676
|
oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor
|
1.00000 |
1.00000 |
1
|
23
|
|
GO:0071813
|
lipoprotein particle binding
|
1.00000 |
1.00000 |
1
|
13
|
|
GO:0004871
|
signal transducer activity
|
1.00000 |
1.00000 |
2
|
559
|