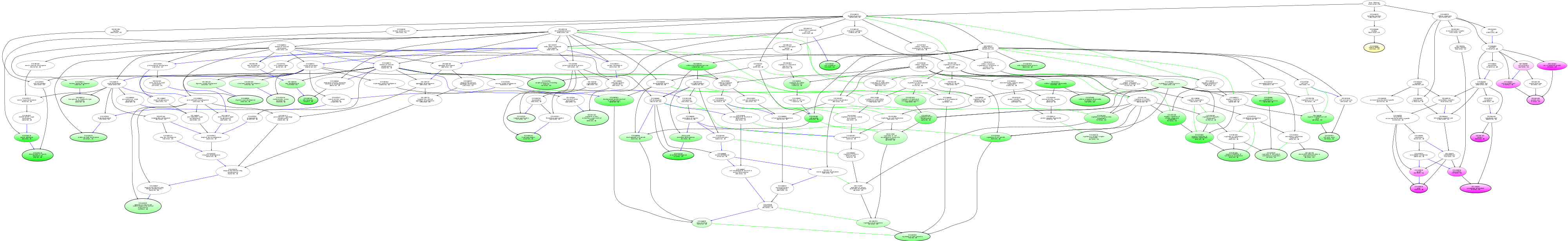
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0030427
|
site of polarized growth
|
0.00386940 |
0.00386940 |
1
|
6
|
|
GO:0005881
|
cytoplasmic microtubule
|
0.00422386 |
0.00422386 |
1
|
8
|
|
GO:0005818
|
aster
|
0.00488998 |
0.00488998 |
1
|
6
|
|
GO:0016204
|
determination of muscle attachment site
|
0.0105263 |
0.0105263 |
1
|
3
|
|
GO:0045169
|
fusome
|
0.0193237 |
0.0193237 |
1
|
36
|
|
GO:0014019
|
neuroblast development
|
0.0207738 |
0.0207738 |
1
|
11
|
|
GO:0016203
|
muscle attachment
|
0.0218696 |
0.0218696 |
1
|
29
|
|
GO:0008037
|
cell recognition
|
0.0221036 |
0.0221036 |
1
|
57
|
|
GO:0022411
|
cellular component disassembly
|
0.0243665 |
0.0243665 |
1
|
50
|
|
GO:0010639
|
negative regulation of organelle organization
|
0.0262515 |
0.0262515 |
1
|
43
|
|
GO:0032886
|
regulation of microtubule-based process
|
0.0271577 |
0.0271577 |
1
|
32
|
|
GO:0016049
|
cell growth
|
0.0274662 |
0.0274662 |
1
|
91
|
|
GO:0035148
|
tube formation
|
0.0278293 |
0.0278293 |
1
|
15
|
|
GO:0051129
|
negative regulation of cellular component organization
|
0.0310688 |
0.0310688 |
1
|
75
|
|
GO:0048869
|
cellular developmental process
|
0.0312311 |
0.0312311 |
2
|
1276
|
|
GO:0005874
|
microtubule
|
0.0315789 |
0.0315789 |
1
|
51
|
|
GO:0071845
|
cellular component disassembly at cellular level
|
0.0329598 |
0.0329598 |
1
|
50
|
|
GO:0060560
|
developmental growth involved in morphogenesis
|
0.0355105 |
0.0355105 |
1
|
56
|
|
GO:0001558
|
regulation of cell growth
|
0.0363519 |
0.0363519 |
1
|
38
|
|
GO:0045786
|
negative regulation of cell cycle
|
0.0369181 |
0.0369181 |
1
|
46
|
|
GO:0044463
|
cell projection part
|
0.0388179 |
0.0388179 |
1
|
61
|
|
GO:0022604
|
regulation of cell morphogenesis
|
0.0388569 |
0.0388569 |
1
|
44
|
|
GO:0035287
|
head segmentation
|
0.0409683 |
0.0409683 |
1
|
22
|
|
GO:0051494
|
negative regulation of cytoskeleton organization
|
0.0456204 |
0.0456204 |
1
|
25
|
|
GO:0030426
|
growth cone
|
0.0480769 |
0.0480769 |
1
|
5
|
|
GO:0031344
|
regulation of cell projection organization
|
0.0498826 |
0.0498826 |
1
|
57
|
|
GO:0007050
|
cell cycle arrest
|
0.0502283 |
0.0502283 |
1
|
33
|
|
GO:0001763
|
morphogenesis of a branching structure
|
0.0517287 |
0.0517287 |
1
|
59
|
|
GO:0005819
|
spindle
|
0.0554197 |
0.0554197 |
1
|
68
|
|
GO:0014016
|
neuroblast differentiation
|
0.0557214 |
0.0557214 |
1
|
35
|
|
GO:0030029
|
actin filament-based process
|
0.0559076 |
0.0559076 |
1
|
186
|
|
GO:0030516
|
regulation of axon extension
|
0.0578512 |
0.0578512 |
1
|
7
|
|
GO:0016319
|
mushroom body development
|
0.0600122 |
0.0600122 |
1
|
69
|
|
GO:0022603
|
regulation of anatomical structure morphogenesis
|
0.0618812 |
0.0618812 |
1
|
100
|
|
GO:0032535
|
regulation of cellular component size
|
0.0631681 |
0.0631681 |
1
|
130
|
|
GO:0002520
|
immune system development
|
0.0644123 |
0.0644123 |
1
|
60
|
|
GO:0048565
|
digestive tract development
|
0.0694077 |
0.0694077 |
1
|
80
|
|
GO:0035239
|
tube morphogenesis
|
0.0759740 |
0.0759740 |
1
|
117
|
|
GO:0048588
|
developmental cell growth
|
0.0764154 |
0.0764154 |
1
|
43
|
|
GO:0031109
|
microtubule polymerization or depolymerization
|
0.0766962 |
0.0766962 |
1
|
26
|
|
GO:0035152
|
regulation of tube architecture, open tracheal system
|
0.0782313 |
0.0782313 |
1
|
69
|
|
GO:0007475
|
apposition of dorsal and ventral imaginal disc-derived wing surfaces
|
0.0800971 |
0.0800971 |
1
|
33
|
|
GO:0070507
|
regulation of microtubule cytoskeleton organization
|
0.0804290 |
0.0804290 |
1
|
30
|
|
GO:0019842
|
vitamin binding
|
0.0855787 |
0.0855787 |
1
|
100
|
|
GO:0010769
|
regulation of cell morphogenesis involved in differentiation
|
0.0862745 |
0.0862745 |
1
|
44
|
|
GO:0050770
|
regulation of axonogenesis
|
0.0868056 |
0.0868056 |
1
|
25
|
|
GO:0072359
|
circulatory system development
|
0.0887662 |
0.0887662 |
1
|
77
|
|
GO:0072358
|
cardiovascular system development
|
0.0887662 |
0.0887662 |
1
|
77
|
|
GO:0033043
|
regulation of organelle organization
|
0.0890453 |
0.0890453 |
1
|
127
|
|
GO:0042995
|
cell projection
|
0.0892053 |
0.0892053 |
1
|
143
|
|
GO:0043244
|
regulation of protein complex disassembly
|
0.0893471 |
0.0893471 |
1
|
26
|
|
GO:0055123
|
digestive system development
|
0.0921411 |
0.0921411 |
1
|
80
|
|
GO:0048675
|
axon extension
|
0.0924092 |
0.0924092 |
1
|
28
|
|
GO:0050794
|
regulation of cellular process
|
0.0938314 |
0.0938314 |
2
|
2034
|
|
GO:0048534
|
hemopoietic or lymphoid organ development
|
0.0943216 |
0.0943216 |
1
|
60
|
|
GO:0007379
|
segment specification
|
0.0949721 |
0.0949721 |
1
|
51
|
|
GO:0035295
|
tube development
|
0.100500 |
0.100500 |
1
|
133
|
|
GO:0005575
|
cellular_component
|
0.109458 |
0.109458 |
6
|
8817
|
|
GO:0008431
|
vitamin E binding
|
0.110000 |
0.110000 |
1
|
11
|
|
GO:0002376
|
immune system process
|
0.110207 |
0.110207 |
1
|
245
|
|
GO:0051493
|
regulation of cytoskeleton organization
|
0.110345 |
0.110345 |
1
|
64
|
|
GO:0040007
|
growth
|
0.111065 |
0.111065 |
1
|
247
|
|
GO:0000235
|
astral microtubule
|
0.111111 |
0.111111 |
1
|
3
|
|
GO:0040008
|
regulation of growth
|
0.111607 |
0.111607 |
1
|
132
|
|
GO:0048589
|
developmental growth
|
0.112049 |
0.112049 |
1
|
154
|
|
GO:0010975
|
regulation of neuron projection development
|
0.114558 |
0.114558 |
1
|
48
|
|
GO:0043933
|
macromolecular complex subunit organization
|
0.116959 |
0.116959 |
1
|
240
|
|
GO:0008038
|
neuron recognition
|
0.118644 |
0.118644 |
1
|
56
|
|
GO:0005488
|
binding
|
0.118821 |
0.118821 |
5
|
5641
|
|
GO:0048638
|
regulation of developmental growth
|
0.118943 |
0.118943 |
1
|
54
|
|
GO:0016198
|
axon choice point recognition
|
0.120000 |
0.120000 |
1
|
24
|
|
GO:0007507
|
heart development
|
0.120197 |
0.120197 |
1
|
77
|
|
GO:0043167
|
ion binding
|
0.120503 |
0.120503 |
3
|
1498
|
|
GO:0045664
|
regulation of neuron differentiation
|
0.131907 |
0.131907 |
1
|
74
|
|
GO:0007017
|
microtubule-based process
|
0.131931 |
0.131931 |
1
|
448
|
|
GO:0007154
|
cell communication
|
0.135620 |
0.135620 |
1
|
461
|
|
GO:0030030
|
cell projection organization
|
0.142264 |
0.142264 |
1
|
485
|
|
GO:0016358
|
dendrite development
|
0.143691 |
0.143691 |
1
|
169
|
|
GO:0051726
|
regulation of cell cycle
|
0.144812 |
0.144812 |
1
|
187
|
|
GO:0048542
|
lymph gland development
|
0.146226 |
0.146226 |
1
|
31
|
|
GO:0032984
|
macromolecular complex disassembly
|
0.151394 |
0.151394 |
1
|
38
|
|
GO:0034621
|
cellular macromolecular complex subunit organization
|
0.152431 |
0.152431 |
1
|
232
|
|
GO:0008587
|
imaginal disc-derived wing margin morphogenesis
|
0.155340 |
0.155340 |
1
|
64
|
|
GO:0034623
|
cellular macromolecular complex disassembly
|
0.156379 |
0.156379 |
1
|
38
|
|
GO:0051128
|
regulation of cellular component organization
|
0.156775 |
0.156775 |
1
|
282
|
|
GO:0050896
|
response to stimulus
|
0.157100 |
0.157100 |
2
|
1548
|
|
GO:0050793
|
regulation of developmental process
|
0.161873 |
0.161873 |
1
|
321
|
|
GO:0035146
|
tube fusion
|
0.162393 |
0.162393 |
1
|
19
|
|
GO:0042221
|
response to chemical stimulus
|
0.166527 |
0.166527 |
2
|
632
|
|
GO:0007383
|
specification of segmental identity, antennal segment
|
0.166667 |
0.166667 |
1
|
3
|
|
GO:0045595
|
regulation of cell differentiation
|
0.167859 |
0.167859 |
1
|
231
|
|
GO:0035149
|
lumen formation, open tracheal system
|
0.173913 |
0.173913 |
1
|
12
|
|
GO:0032501
|
multicellular organismal process
|
0.179631 |
0.179631 |
3
|
3315
|
|
GO:0050890
|
cognition
|
0.181675 |
0.181675 |
1
|
115
|
|
GO:0048523
|
negative regulation of cellular process
|
0.183975 |
0.183975 |
1
|
635
|
|
GO:0035107
|
appendage morphogenesis
|
0.184365 |
0.184365 |
1
|
283
|
|
GO:0007420
|
brain development
|
0.188384 |
0.188384 |
1
|
127
|
|
GO:0007267
|
cell-cell signaling
|
0.188801 |
0.188801 |
1
|
263
|
|
GO:0030716
|
oocyte fate determination
|
0.189394 |
0.189394 |
1
|
25
|
|
GO:0022402
|
cell cycle process
|
0.189739 |
0.189739 |
1
|
655
|
|
GO:0061138
|
morphogenesis of a branching epithelium
|
0.192440 |
0.192440 |
1
|
56
|
|
GO:0048729
|
tissue morphogenesis
|
0.194151 |
0.194151 |
1
|
312
|
|
GO:0001071
|
nucleic acid binding transcription factor activity
|
0.196016 |
0.196016 |
1
|
395
|
|
GO:0001709
|
cell fate determination
|
0.196268 |
0.196268 |
1
|
132
|
|
GO:0022008
|
neurogenesis
|
0.204486 |
0.204486 |
2
|
632
|
|
GO:0007611
|
learning or memory
|
0.205725 |
0.205725 |
1
|
115
|
|
GO:2000026
|
regulation of multicellular organismal development
|
0.207572 |
0.207572 |
1
|
259
|
|
GO:0040011
|
locomotion
|
0.208134 |
0.208134 |
1
|
484
|
|
GO:0060446
|
branching involved in open tracheal system development
|
0.208178 |
0.208178 |
1
|
56
|
|
GO:0048468
|
cell development
|
0.209123 |
0.209123 |
2
|
1042
|
|
GO:0048736
|
appendage development
|
0.209532 |
0.209532 |
1
|
286
|
|
GO:0061061
|
muscle structure development
|
0.210182 |
0.210182 |
1
|
252
|
|
GO:0030528
|
transcription regulator activity
|
0.212605 |
0.212605 |
1
|
432
|
|
GO:0007049
|
cell cycle
|
0.217222 |
0.217222 |
1
|
756
|
|
GO:0005876
|
spindle microtubule
|
0.221053 |
0.221053 |
1
|
21
|
|
GO:0044430
|
cytoskeletal part
|
0.231674 |
0.231674 |
1
|
512
|
|
GO:0090066
|
regulation of anatomical structure size
|
0.231824 |
0.231824 |
1
|
169
|
|
GO:0051239
|
regulation of multicellular organismal process
|
0.232587 |
0.232587 |
1
|
368
|
|
GO:0044459
|
plasma membrane part
|
0.236675 |
0.236675 |
1
|
373
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.237215 |
0.237215 |
1
|
1039
|
|
GO:0007380
|
specification of segmental identity, head
|
0.237288 |
0.237288 |
1
|
14
|
|
GO:0008092
|
cytoskeletal protein binding
|
0.238743 |
0.238743 |
1
|
220
|
|
GO:0050767
|
regulation of neurogenesis
|
0.245013 |
0.245013 |
1
|
90
|
|
GO:0051960
|
regulation of nervous system development
|
0.247039 |
0.247039 |
1
|
124
|
|
GO:0007399
|
nervous system development
|
0.249853 |
0.249853 |
2
|
848
|
|
GO:0007417
|
central nervous system development
|
0.252905 |
0.252905 |
1
|
230
|
|
GO:0043242
|
negative regulation of protein complex disassembly
|
0.258427 |
0.258427 |
1
|
23
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.259446 |
0.259446 |
1
|
412
|
|
GO:0043241
|
protein complex disassembly
|
0.268116 |
0.268116 |
1
|
37
|
|
GO:0030054
|
cell junction
|
0.270777 |
0.270777 |
1
|
101
|
|
GO:0060541
|
respiratory system development
|
0.272151 |
0.272151 |
1
|
249
|
|
GO:0007552
|
metamorphosis
|
0.273794 |
0.273794 |
1
|
420
|
|
GO:0007517
|
muscle organ development
|
0.276072 |
0.276072 |
1
|
191
|
|
GO:0060562
|
epithelial tube morphogenesis
|
0.281150 |
0.281150 |
1
|
88
|
|
GO:0006357
|
regulation of transcription from RNA polymerase II promoter
|
0.282934 |
0.282934 |
1
|
189
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.283034 |
0.283034 |
1
|
1056
|
|
GO:0048732
|
gland development
|
0.284024 |
0.284024 |
1
|
191
|
|
GO:0050789
|
regulation of biological process
|
0.286552 |
0.286552 |
2
|
2246
|
|
GO:0032774
|
RNA biosynthetic process
|
0.290017 |
0.290017 |
1
|
703
|
|
GO:0048519
|
negative regulation of biological process
|
0.291182 |
0.291182 |
1
|
706
|
|
GO:0005886
|
plasma membrane
|
0.292027 |
0.292027 |
1
|
641
|
|
GO:0003704
|
specific RNA polymerase II transcription factor activity
|
0.292135 |
0.292135 |
1
|
78
|
|
GO:0060284
|
regulation of cell development
|
0.292999 |
0.292999 |
1
|
168
|
|
GO:0009880
|
embryonic pattern specification
|
0.294591 |
0.294591 |
1
|
256
|
|
GO:0048513
|
organ development
|
0.300572 |
0.300572 |
2
|
1242
|
|
GO:0043565
|
sequence-specific DNA binding
|
0.300971 |
0.300971 |
1
|
248
|
|
GO:0015631
|
tubulin binding
|
0.309091 |
0.309091 |
1
|
68
|
|
GO:0048646
|
anatomical structure formation involved in morphogenesis
|
0.322148 |
0.322148 |
1
|
466
|
|
GO:0007635
|
chemosensory behavior
|
0.338282 |
0.338282 |
1
|
176
|
|
GO:0035147
|
branch fusion, open tracheal system
|
0.339286 |
0.339286 |
1
|
19
|
|
GO:0030036
|
actin cytoskeleton organization
|
0.339518 |
0.339518 |
1
|
183
|
|
GO:0005622
|
intracellular
|
0.340893 |
0.340893 |
4
|
4734
|
|
GO:0009791
|
post-embryonic development
|
0.343209 |
0.343209 |
1
|
500
|
|
GO:0065007
|
biological regulation
|
0.346129 |
0.346129 |
2
|
2548
|
|
GO:0032989
|
cellular component morphogenesis
|
0.348979 |
0.348979 |
1
|
566
|
|
GO:0007494
|
midgut development
|
0.350000 |
0.350000 |
1
|
28
|
|
GO:0045165
|
cell fate commitment
|
0.359875 |
0.359875 |
1
|
255
|
|
GO:0006366
|
transcription from RNA polymerase II promoter
|
0.360913 |
0.360913 |
1
|
253
|
|
GO:0032502
|
developmental process
|
0.364008 |
0.364008 |
2
|
2638
|
|
GO:0007389
|
pattern specification process
|
0.365750 |
0.365750 |
1
|
537
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.366791 |
0.366791 |
1
|
888
|
|
GO:0005509
|
calcium ion binding
|
0.373519 |
0.373519 |
1
|
209
|
|
GO:0019222
|
regulation of metabolic process
|
0.376562 |
0.376562 |
1
|
1242
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.381695 |
0.381695 |
1
|
905
|
|
GO:0005215
|
transporter activity
|
0.381781 |
0.381781 |
1
|
852
|
|
GO:0006350
|
transcription
|
0.387810 |
0.387810 |
1
|
859
|
|
GO:0042330
|
taxis
|
0.389803 |
0.389803 |
1
|
237
|
|
GO:0071944
|
cell periphery
|
0.391801 |
0.391801 |
1
|
724
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.396252 |
0.396252 |
1
|
888
|
|
GO:0009058
|
biosynthetic process
|
0.402387 |
0.402387 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.402965 |
0.402965 |
1
|
2093
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.405649 |
0.405649 |
1
|
1623
|
|
GO:0035288
|
anterior head segmentation
|
0.409091 |
0.409091 |
1
|
9
|
|
GO:0016070
|
RNA metabolic process
|
0.413829 |
0.413829 |
1
|
1191
|
|
GO:0048563
|
post-embryonic organ morphogenesis
|
0.415167 |
0.415167 |
1
|
323
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.415756 |
0.415756 |
1
|
1789
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.418447 |
0.418447 |
1
|
1562
|
|
GO:0048813
|
dendrite morphogenesis
|
0.419192 |
0.419192 |
1
|
166
|
|
GO:0009605
|
response to external stimulus
|
0.425931 |
0.425931 |
1
|
375
|
|
GO:0009790
|
embryo development
|
0.427001 |
0.427001 |
1
|
641
|
|
GO:0003674
|
molecular_function
|
0.427511 |
0.427511 |
6
|
11064
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.429576 |
0.429576 |
1
|
1110
|
|
GO:0009888
|
tissue development
|
0.431439 |
0.431439 |
1
|
557
|
|
GO:0048731
|
system development
|
0.432376 |
0.432376 |
2
|
1696
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.440308 |
0.440308 |
1
|
686
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.451726 |
0.451726 |
1
|
903
|
|
GO:0007010
|
cytoskeleton organization
|
0.453469 |
0.453469 |
1
|
536
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.456989 |
0.456989 |
1
|
850
|
|
GO:0005856
|
cytoskeleton
|
0.457213 |
0.457213 |
1
|
561
|
|
GO:0048754
|
branching morphogenesis of a tube
|
0.466667 |
0.466667 |
1
|
56
|
|
GO:0048569
|
post-embryonic organ development
|
0.476228 |
0.476228 |
1
|
343
|
|
GO:0008355
|
olfactory learning
|
0.481250 |
0.481250 |
1
|
77
|
|
GO:0005515
|
protein binding
|
0.484079 |
0.484079 |
2
|
1726
|
|
GO:0003008
|
system process
|
0.488695 |
0.488695 |
1
|
664
|
|
GO:0010468
|
regulation of gene expression
|
0.489930 |
0.489930 |
1
|
973
|
|
GO:0065008
|
regulation of biological quality
|
0.490437 |
0.490437 |
1
|
729
|
|
GO:0006935
|
chemotaxis
|
0.492835 |
0.492835 |
1
|
193
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.496927 |
0.496927 |
1
|
1617
|
|
GO:0023052
|
signaling
|
0.497295 |
0.497295 |
1
|
1364
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.498346 |
0.498346 |
1
|
1959
|
|
GO:0043005
|
neuron projection
|
0.503497 |
0.503497 |
1
|
72
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.513595 |
0.513595 |
1
|
850
|
|
GO:0044249
|
cellular biosynthetic process
|
0.526807 |
0.526807 |
1
|
2034
|
|
GO:0060429
|
epithelium development
|
0.529623 |
0.529623 |
1
|
295
|
|
GO:0000226
|
microtubule cytoskeleton organization
|
0.529687 |
0.529687 |
1
|
339
|
|
GO:0003676
|
nucleic acid binding
|
0.530250 |
0.530250 |
2
|
1855
|
|
GO:0010467
|
gene expression
|
0.531494 |
0.531494 |
1
|
1907
|
|
GO:0060538
|
skeletal muscle organ development
|
0.539267 |
0.539267 |
1
|
103
|
|
GO:0003779
|
actin binding
|
0.545455 |
0.545455 |
1
|
120
|
|
GO:0051234
|
establishment of localization
|
0.545594 |
0.545594 |
1
|
1549
|
|
GO:0070161
|
anchoring junction
|
0.564356 |
0.564356 |
1
|
57
|
|
GO:0071822
|
protein complex subunit organization
|
0.570833 |
0.570833 |
1
|
137
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.573530 |
0.573530 |
1
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.574428 |
0.574428 |
1
|
1227
|
|
GO:0007560
|
imaginal disc morphogenesis
|
0.575758 |
0.575758 |
1
|
323
|
|
GO:0035282
|
segmentation
|
0.581028 |
0.581028 |
1
|
294
|
|
GO:0007610
|
behavior
|
0.591970 |
0.591970 |
1
|
559
|
|
GO:0009887
|
organ morphogenesis
|
0.608733 |
0.608733 |
1
|
684
|
|
GO:0008270
|
zinc ion binding
|
0.610500 |
0.610500 |
2
|
901
|
|
GO:0007444
|
imaginal disc development
|
0.610821 |
0.610821 |
1
|
467
|
|
GO:0000904
|
cell morphogenesis involved in differentiation
|
0.611753 |
0.611753 |
1
|
408
|
|
GO:0003702
|
RNA polymerase II transcription factor activity
|
0.618056 |
0.618056 |
1
|
267
|
|
GO:0051179
|
localization
|
0.626157 |
0.626157 |
1
|
1896
|
|
GO:0035120
|
post-embryonic appendage morphogenesis
|
0.638095 |
0.638095 |
1
|
268
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.639129 |
0.639129 |
1
|
2878
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0044425
|
membrane part
|
0.640589 |
0.640589 |
1
|
1398
|
|
GO:0043229
|
intracellular organelle
|
0.646541 |
0.646541 |
2
|
3530
|
|
GO:0007472
|
wing disc morphogenesis
|
0.665796 |
0.665796 |
1
|
255
|
|
GO:0044444
|
cytoplasmic part
|
0.669378 |
0.669378 |
1
|
1863
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.669454 |
0.669454 |
1
|
2108
|
|
GO:0007409
|
axonogenesis
|
0.679389 |
0.679389 |
1
|
267
|
|
GO:0048666
|
neuron development
|
0.679533 |
0.679533 |
1
|
471
|
|
GO:0035220
|
wing disc development
|
0.680942 |
0.680942 |
1
|
318
|
|
GO:0007411
|
axon guidance
|
0.683824 |
0.683824 |
1
|
186
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.684327 |
0.684327 |
1
|
620
|
|
GO:0007423
|
sensory organ development
|
0.684358 |
0.684358 |
1
|
544
|
|
GO:0045449
|
regulation of transcription
|
0.687166 |
0.687166 |
1
|
771
|
|
GO:0030182
|
neuron differentiation
|
0.689161 |
0.689161 |
1
|
548
|
|
GO:0043170
|
macromolecule metabolic process
|
0.690797 |
0.690797 |
1
|
3588
|
|
GO:0003677
|
DNA binding
|
0.691225 |
0.691225 |
1
|
824
|
|
GO:0031175
|
neuron projection development
|
0.696270 |
0.696270 |
1
|
392
|
|
GO:0007019
|
microtubule depolymerization
|
0.696970 |
0.696970 |
1
|
23
|
|
GO:0031111
|
negative regulation of microtubule polymerization or depolymerization
|
0.700000 |
0.700000 |
1
|
21
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.724105 |
0.724105 |
1
|
1517
|
|
GO:0043234
|
protein complex
|
0.727104 |
0.727104 |
1
|
1564
|
|
GO:0007612
|
learning
|
0.730435 |
0.730435 |
1
|
84
|
|
GO:0008150
|
biological_process
|
0.735561 |
0.735561 |
5
|
10616
|
|
GO:0048856
|
anatomical structure development
|
0.737157 |
0.737157 |
2
|
2265
|
|
GO:0007275
|
multicellular organismal development
|
0.738485 |
0.738485 |
2
|
2296
|
|
GO:0044237
|
cellular metabolic process
|
0.740940 |
0.740940 |
1
|
3814
|
|
GO:0044446
|
intracellular organelle part
|
0.743053 |
0.743053 |
1
|
2162
|
|
GO:0032990
|
cell part morphogenesis
|
0.745583 |
0.745583 |
1
|
422
|
|
GO:0005623
|
cell
|
0.749201 |
0.749201 |
4
|
6195
|
|
GO:0044464
|
cell part
|
0.749201 |
0.749201 |
4
|
6195
|
|
GO:0031110
|
regulation of microtubule polymerization or depolymerization
|
0.750000 |
0.750000 |
1
|
24
|
|
GO:0043226
|
organelle
|
0.768603 |
0.768603 |
2
|
3537
|
|
GO:0008361
|
regulation of cell size
|
0.769231 |
0.769231 |
1
|
100
|
|
GO:0006996
|
organelle organization
|
0.779169 |
0.779169 |
1
|
1182
|
|
GO:0007350
|
blastoderm segmentation
|
0.779221 |
0.779221 |
1
|
240
|
|
GO:0005737
|
cytoplasm
|
0.790684 |
0.790684 |
1
|
2378
|
|
GO:0016199
|
axon midline choice point recognition
|
0.791667 |
0.791667 |
1
|
19
|
|
GO:0048667
|
cell morphogenesis involved in neuron differentiation
|
0.795501 |
0.795501 |
1
|
389
|
|
GO:0015630
|
microtubule cytoskeleton
|
0.798574 |
0.798574 |
1
|
448
|
|
GO:0048858
|
cell projection morphogenesis
|
0.806883 |
0.806883 |
1
|
422
|
|
GO:0051261
|
protein depolymerization
|
0.810811 |
0.810811 |
1
|
30
|
|
GO:0016020
|
membrane
|
0.813245 |
0.813245 |
1
|
2122
|
|
GO:0032991
|
macromolecular complex
|
0.813350 |
0.813350 |
1
|
2151
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.814170 |
0.814170 |
1
|
701
|
|
GO:0044422
|
organelle part
|
0.817512 |
0.817512 |
1
|
2176
|
|
GO:0044238
|
primary metabolic process
|
0.819985 |
0.819985 |
1
|
4259
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.824951 |
0.824951 |
1
|
1534
|
|
GO:0000902
|
cell morphogenesis
|
0.825088 |
0.825088 |
1
|
467
|
|
GO:0007026
|
negative regulation of microtubule depolymerization
|
0.840000 |
0.840000 |
1
|
21
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.842767 |
0.842767 |
1
|
402
|
|
GO:0031114
|
regulation of microtubule depolymerization
|
0.851852 |
0.851852 |
1
|
23
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0042048
|
olfactory behavior
|
0.892045 |
0.892045 |
1
|
157
|
|
GO:0046914
|
transition metal ion binding
|
0.892059 |
0.892059 |
2
|
1153
|
|
GO:0007476
|
imaginal disc-derived wing morphogenesis
|
0.903915 |
0.903915 |
1
|
254
|
|
GO:0046872
|
metal ion binding
|
0.906799 |
0.906799 |
3
|
1449
|
|
GO:0002009
|
morphogenesis of an epithelium
|
0.908805 |
0.908805 |
1
|
289
|
|
GO:0048699
|
generation of neurons
|
0.910264 |
0.910264 |
2
|
603
|
|
GO:0048812
|
neuron projection morphogenesis
|
0.910798 |
0.910798 |
1
|
388
|
|
GO:0044424
|
intracellular part
|
0.922046 |
0.922046 |
2
|
4384
|
|
GO:0009987
|
cellular process
|
0.926071 |
0.926071 |
2
|
6560
|
|
GO:0008017
|
microtubule binding
|
0.941176 |
0.941176 |
1
|
64
|
|
GO:0003002
|
regionalization
|
0.942272 |
0.942272 |
1
|
506
|
|
GO:0030154
|
cell differentiation
|
0.942825 |
0.942825 |
2
|
1239
|
|
GO:0008306
|
associative learning
|
0.952381 |
0.952381 |
1
|
80
|
|
GO:0050877
|
neurological system process
|
0.953313 |
0.953313 |
1
|
633
|
|
GO:0002165
|
instar larval or pupal development
|
0.954000 |
0.954000 |
1
|
477
|
|
GO:0043227
|
membrane-bounded organelle
|
0.963300 |
0.963300 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.963413 |
0.963413 |
1
|
2856
|
|
GO:0008152
|
metabolic process
|
0.965278 |
0.965278 |
1
|
5194
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0043624
|
cellular protein complex disassembly
|
0.973684 |
0.973684 |
1
|
37
|
|
GO:0035114
|
imaginal disc-derived appendage morphogenesis
|
0.975524 |
0.975524 |
1
|
279
|
|
GO:0005912
|
adherens junction
|
0.982456 |
0.982456 |
1
|
56
|
|
GO:0048737
|
imaginal disc-derived appendage development
|
0.986014 |
0.986014 |
1
|
282
|
|
GO:0007424
|
open tracheal system development
|
0.995984 |
0.995984 |
1
|
248
|
|
GO:0043169
|
cation binding
|
0.997997 |
0.997997 |
3
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
6
|
12747
|
|
GO:0003700
|
sequence-specific DNA binding transcription factor activity
|
1.00000 |
1.00000 |
1
|
395
|