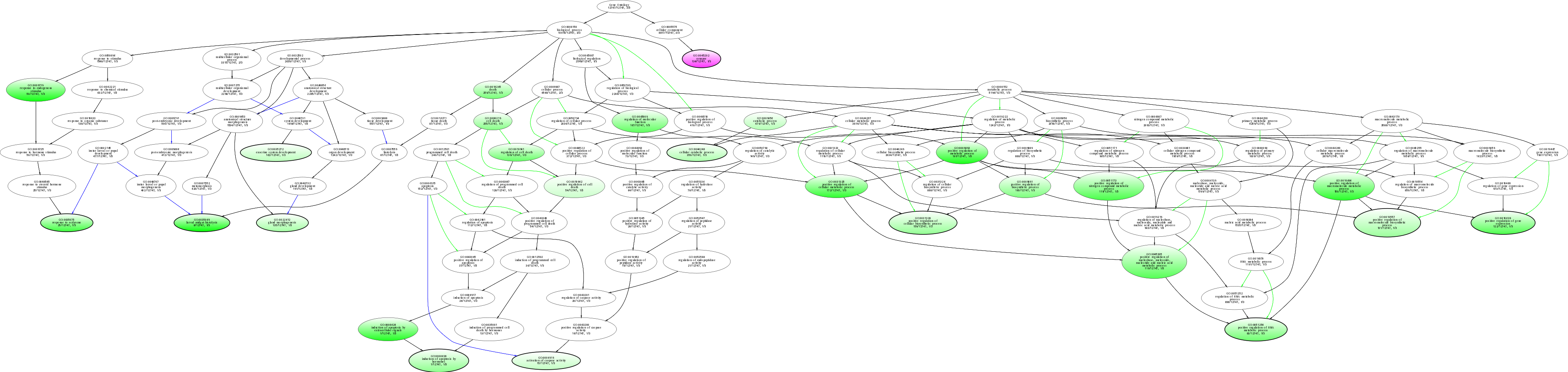
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0035069
|
larval midgut histolysis
|
0.0190476 |
0.0190476 |
1
|
8
|
|
GO:0009719
|
response to endogenous stimulus
|
0.0322997 |
0.0322997 |
1
|
50
|
|
GO:0009893
|
positive regulation of metabolic process
|
0.0341381 |
0.0341381 |
1
|
181
|
|
GO:0008624
|
induction of apoptosis by extracellular signals
|
0.0384615 |
0.0384615 |
1
|
1
|
|
GO:0010604
|
positive regulation of macromolecule metabolic process
|
0.0415858 |
0.0415858 |
1
|
150
|
|
GO:0031325
|
positive regulation of cellular metabolic process
|
0.0438776 |
0.0438776 |
1
|
172
|
|
GO:0045202
|
synapse
|
0.0449093 |
0.0449093 |
1
|
134
|
|
GO:0035075
|
response to ecdysone
|
0.0524109 |
0.0524109 |
1
|
25
|
|
GO:0051173
|
positive regulation of nitrogen compound metabolic process
|
0.0557638 |
0.0557638 |
1
|
119
|
|
GO:0010628
|
positive regulation of gene expression
|
0.0636411 |
0.0636411 |
1
|
122
|
|
GO:0045935
|
positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.0649918 |
0.0649918 |
1
|
119
|
|
GO:0065009
|
regulation of molecular function
|
0.0655416 |
0.0655416 |
1
|
167
|
|
GO:0010941
|
regulation of cell death
|
0.0658768 |
0.0658768 |
1
|
139
|
|
GO:0051254
|
positive regulation of RNA metabolic process
|
0.0676998 |
0.0676998 |
1
|
83
|
|
GO:0016265
|
death
|
0.0714268 |
0.0714268 |
1
|
259
|
|
GO:0009891
|
positive regulation of biosynthetic process
|
0.0760818 |
0.0760818 |
1
|
160
|
|
GO:0008219
|
cell death
|
0.0762386 |
0.0762386 |
1
|
255
|
|
GO:0008628
|
induction of apoptosis by hormones
|
0.0769231 |
0.0769231 |
1
|
1
|
|
GO:0031328
|
positive regulation of cellular biosynthetic process
|
0.0777886 |
0.0777886 |
1
|
159
|
|
GO:0010557
|
positive regulation of macromolecule biosynthetic process
|
0.0789632 |
0.0789632 |
1
|
131
|
|
GO:0022612
|
gland morphogenesis
|
0.0873786 |
0.0873786 |
1
|
135
|
|
GO:0044248
|
cellular catabolic process
|
0.0902527 |
0.0902527 |
1
|
350
|
|
GO:0009056
|
catabolic process
|
0.0922218 |
0.0922218 |
1
|
479
|
|
GO:0035272
|
exocrine system development
|
0.0955189 |
0.0955189 |
1
|
162
|
|
GO:0006919
|
activation of caspase activity
|
0.0986842 |
0.0986842 |
1
|
15
|
|
GO:0010942
|
positive regulation of cell death
|
0.0996310 |
0.0996310 |
1
|
54
|
|
GO:0016044
|
cellular membrane organization
|
0.102136 |
0.102136 |
1
|
344
|
|
GO:0001071
|
nucleic acid binding transcription factor activity
|
0.103335 |
0.103335 |
1
|
395
|
|
GO:0048522
|
positive regulation of cellular process
|
0.110028 |
0.110028 |
1
|
372
|
|
GO:0048518
|
positive regulation of biological process
|
0.111456 |
0.111456 |
1
|
410
|
|
GO:0045893
|
positive regulation of transcription, DNA-dependent
|
0.114685 |
0.114685 |
1
|
82
|
|
GO:0006914
|
autophagy
|
0.117143 |
0.117143 |
1
|
41
|
|
GO:0050790
|
regulation of catalytic activity
|
0.119487 |
0.119487 |
1
|
149
|
|
GO:0016192
|
vesicle-mediated transport
|
0.121581 |
0.121581 |
1
|
430
|
|
GO:0045941
|
positive regulation of transcription
|
0.127636 |
0.127636 |
1
|
115
|
|
GO:0007154
|
cell communication
|
0.135620 |
0.135620 |
1
|
461
|
|
GO:0035072
|
ecdysone-mediated induction of salivary gland cell autophagic cell death
|
0.142857 |
0.142857 |
1
|
11
|
|
GO:0048732
|
gland development
|
0.153784 |
0.153784 |
1
|
191
|
|
GO:0007559
|
histolysis
|
0.156194 |
0.156194 |
1
|
87
|
|
GO:0061024
|
membrane organization
|
0.168129 |
0.168129 |
1
|
345
|
|
GO:0035070
|
salivary gland histolysis
|
0.175325 |
0.175325 |
1
|
81
|
|
GO:0043281
|
regulation of caspase activity
|
0.176991 |
0.176991 |
1
|
20
|
|
GO:0007267
|
cell-cell signaling
|
0.188801 |
0.188801 |
1
|
263
|
|
GO:0009791
|
post-embryonic development
|
0.189538 |
0.189538 |
1
|
500
|
|
GO:0019226
|
transmission of nerve impulse
|
0.194826 |
0.194826 |
1
|
241
|
|
GO:0023046
|
signaling process
|
0.199779 |
0.199779 |
1
|
760
|
|
GO:0019222
|
regulation of metabolic process
|
0.210401 |
0.210401 |
1
|
1242
|
|
GO:0010033
|
response to organic substance
|
0.218354 |
0.218354 |
1
|
138
|
|
GO:0043068
|
positive regulation of programmed cell death
|
0.225000 |
0.225000 |
1
|
54
|
|
GO:0043065
|
positive regulation of apoptosis
|
0.231250 |
0.231250 |
1
|
37
|
|
GO:0032501
|
multicellular organismal process
|
0.231607 |
0.231607 |
2
|
3315
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.237215 |
0.237215 |
1
|
1039
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.244709 |
0.244709 |
1
|
1110
|
|
GO:0051345
|
positive regulation of hydrolase activity
|
0.245614 |
0.245614 |
1
|
28
|
|
GO:0009888
|
tissue development
|
0.245916 |
0.245916 |
1
|
557
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.259446 |
0.259446 |
1
|
412
|
|
GO:0052547
|
regulation of peptidase activity
|
0.269231 |
0.269231 |
1
|
21
|
|
GO:0007552
|
metamorphosis
|
0.273794 |
0.273794 |
1
|
420
|
|
GO:0005700
|
polytene chromosome
|
0.276650 |
0.276650 |
1
|
109
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.283034 |
0.283034 |
1
|
1056
|
|
GO:0032774
|
RNA biosynthetic process
|
0.290017 |
0.290017 |
1
|
703
|
|
GO:0048102
|
autophagic cell death
|
0.316667 |
0.316667 |
1
|
76
|
|
GO:0005694
|
chromosome
|
0.321108 |
0.321108 |
1
|
394
|
|
GO:0005575
|
cellular_component
|
0.330897 |
0.330897 |
3
|
8817
|
|
GO:0016271
|
tissue death
|
0.335907 |
0.335907 |
1
|
87
|
|
GO:0023052
|
signaling
|
0.338079 |
0.338079 |
1
|
1364
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.346904 |
0.346904 |
1
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.347592 |
0.347592 |
1
|
1227
|
|
GO:0003008
|
system process
|
0.360531 |
0.360531 |
1
|
664
|
|
GO:0035081
|
induction of programmed cell death by hormones
|
0.361111 |
0.361111 |
1
|
13
|
|
GO:0009725
|
response to hormone stimulus
|
0.362319 |
0.362319 |
1
|
50
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.366791 |
0.366791 |
1
|
888
|
|
GO:0050896
|
response to stimulus
|
0.376795 |
0.376795 |
1
|
1548
|
|
GO:0051234
|
establishment of localization
|
0.377001 |
0.377001 |
1
|
1549
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.381695 |
0.381695 |
1
|
905
|
|
GO:0006350
|
transcription
|
0.387810 |
0.387810 |
1
|
859
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.396252 |
0.396252 |
1
|
888
|
|
GO:0009058
|
biosynthetic process
|
0.402387 |
0.402387 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.402965 |
0.402965 |
1
|
2093
|
|
GO:0043085
|
positive regulation of catalytic activity
|
0.405063 |
0.405063 |
1
|
64
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.405649 |
0.405649 |
1
|
1623
|
|
GO:0042221
|
response to chemical stimulus
|
0.408269 |
0.408269 |
1
|
632
|
|
GO:0016070
|
RNA metabolic process
|
0.413829 |
0.413829 |
1
|
1191
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.415756 |
0.415756 |
1
|
1789
|
|
GO:0044093
|
positive regulation of molecular function
|
0.437126 |
0.437126 |
1
|
73
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.440308 |
0.440308 |
1
|
686
|
|
GO:0003677
|
DNA binding
|
0.444205 |
0.444205 |
1
|
824
|
|
GO:0051179
|
localization
|
0.445834 |
0.445834 |
1
|
1896
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.451726 |
0.451726 |
1
|
903
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.456989 |
0.456989 |
1
|
850
|
|
GO:0035078
|
induction of programmed cell death by ecdysone
|
0.461538 |
0.461538 |
1
|
12
|
|
GO:0010952
|
positive regulation of peptidase activity
|
0.484848 |
0.484848 |
1
|
16
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.485282 |
0.485282 |
1
|
2108
|
|
GO:0010468
|
regulation of gene expression
|
0.489930 |
0.489930 |
1
|
973
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.496927 |
0.496927 |
1
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.498346 |
0.498346 |
1
|
1959
|
|
GO:0048545
|
response to steroid hormone stimulus
|
0.500000 |
0.500000 |
1
|
25
|
|
GO:0050789
|
regulation of biological process
|
0.509927 |
0.509927 |
1
|
2246
|
|
GO:0043067
|
regulation of programmed cell death
|
0.509960 |
0.509960 |
1
|
128
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.513595 |
0.513595 |
1
|
850
|
|
GO:0005488
|
binding
|
0.514777 |
0.514777 |
2
|
5641
|
|
GO:0050794
|
regulation of cellular process
|
0.518911 |
0.518911 |
1
|
2034
|
|
GO:0051336
|
regulation of hydrolase activity
|
0.523490 |
0.523490 |
1
|
78
|
|
GO:0044249
|
cellular biosynthetic process
|
0.526807 |
0.526807 |
1
|
2034
|
|
GO:0010467
|
gene expression
|
0.531494 |
0.531494 |
1
|
1907
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0003676
|
nucleic acid binding
|
0.549587 |
0.549587 |
1
|
1855
|
|
GO:0065007
|
biological regulation
|
0.561089 |
0.561089 |
1
|
2548
|
|
GO:0006917
|
induction of apoptosis
|
0.565217 |
0.565217 |
1
|
26
|
|
GO:0032502
|
developmental process
|
0.575616 |
0.575616 |
1
|
2638
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0006915
|
apoptosis
|
0.633333 |
0.633333 |
1
|
152
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.639129 |
0.639129 |
1
|
2878
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0006897
|
endocytosis
|
0.655814 |
0.655814 |
1
|
282
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0012502
|
induction of programmed cell death
|
0.666667 |
0.666667 |
1
|
36
|
|
GO:0009987
|
cellular process
|
0.673638 |
0.673638 |
2
|
6560
|
|
GO:0042981
|
regulation of apoptosis
|
0.674699 |
0.674699 |
1
|
112
|
|
GO:0006911
|
phagocytosis, engulfment
|
0.680851 |
0.680851 |
1
|
192
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.684327 |
0.684327 |
1
|
620
|
|
GO:0045449
|
regulation of transcription
|
0.687166 |
0.687166 |
1
|
771
|
|
GO:0043170
|
macromolecule metabolic process
|
0.690797 |
0.690797 |
1
|
3588
|
|
GO:0044424
|
intracellular part
|
0.707667 |
0.707667 |
1
|
4384
|
|
GO:0006909
|
phagocytosis
|
0.723404 |
0.723404 |
1
|
204
|
|
GO:0044237
|
cellular metabolic process
|
0.740940 |
0.740940 |
1
|
3814
|
|
GO:0005622
|
intracellular
|
0.764165 |
0.764165 |
1
|
4734
|
|
GO:0043226
|
organelle
|
0.785296 |
0.785296 |
1
|
3537
|
|
GO:0043280
|
positive regulation of caspase activity
|
0.800000 |
0.800000 |
1
|
16
|
|
GO:0043229
|
intracellular organelle
|
0.804100 |
0.804100 |
1
|
3530
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.814170 |
0.814170 |
1
|
701
|
|
GO:0010324
|
membrane invagination
|
0.819767 |
0.819767 |
1
|
282
|
|
GO:0044238
|
primary metabolic process
|
0.819985 |
0.819985 |
1
|
4259
|
|
GO:0007435
|
salivary gland morphogenesis
|
0.833333 |
0.833333 |
1
|
135
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.842767 |
0.842767 |
1
|
402
|
|
GO:0007431
|
salivary gland development
|
0.848168 |
0.848168 |
1
|
162
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0007268
|
synaptic transmission
|
0.866667 |
0.866667 |
1
|
234
|
|
GO:0008152
|
metabolic process
|
0.866808 |
0.866808 |
1
|
5194
|
|
GO:0007275
|
multicellular organismal development
|
0.887792 |
0.887792 |
1
|
2296
|
|
GO:0035071
|
salivary gland cell autophagic cell death
|
0.938272 |
0.938272 |
1
|
76
|
|
GO:0012501
|
programmed cell death
|
0.941176 |
0.941176 |
1
|
240
|
|
GO:0050877
|
neurological system process
|
0.953313 |
0.953313 |
1
|
633
|
|
GO:0002165
|
instar larval or pupal development
|
0.954000 |
0.954000 |
1
|
477
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0005623
|
cell
|
0.973722 |
0.973722 |
1
|
6195
|
|
GO:0044464
|
cell part
|
0.973722 |
0.973722 |
1
|
6195
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0052548
|
regulation of endopeptidase activity
|
1.00000 |
1.00000 |
1
|
21
|
|
GO:0003700
|
sequence-specific DNA binding transcription factor activity
|
1.00000 |
1.00000 |
1
|
395
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|