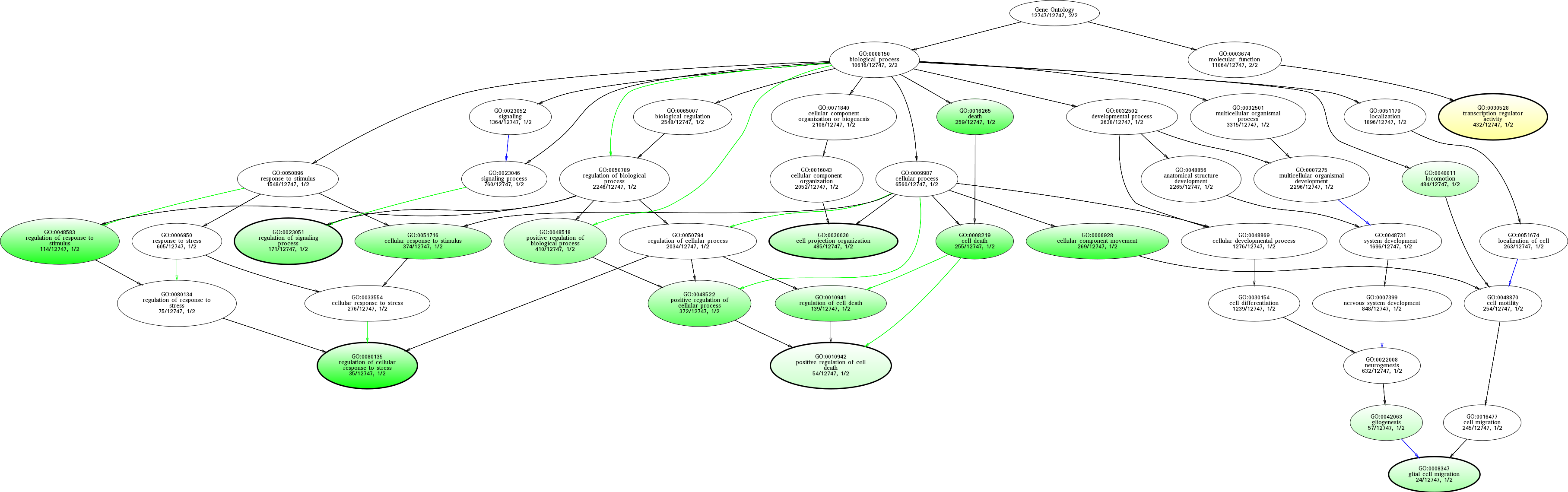
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0080135
|
regulation of cellular response to stress
|
0.0160920 |
0.0160920 |
1
|
35
|
|
GO:0048583
|
regulation of response to stimulus
|
0.0356250 |
0.0356250 |
1
|
114
|
|
GO:0008219
|
cell death
|
0.0388720 |
0.0388720 |
1
|
255
|
|
GO:0006928
|
cellular component movement
|
0.0410061 |
0.0410061 |
1
|
269
|
|
GO:0016265
|
death
|
0.0482013 |
0.0482013 |
1
|
259
|
|
GO:0051716
|
cellular response to stimulus
|
0.0527950 |
0.0527950 |
1
|
374
|
|
GO:0048522
|
positive regulation of cellular process
|
0.0566124 |
0.0566124 |
1
|
372
|
|
GO:0010941
|
regulation of cell death
|
0.0658768 |
0.0658768 |
1
|
139
|
|
GO:0023051
|
regulation of signaling process
|
0.0723656 |
0.0723656 |
1
|
171
|
|
GO:0030030
|
cell projection organization
|
0.0738541 |
0.0738541 |
1
|
485
|
|
GO:0048518
|
positive regulation of biological process
|
0.0757538 |
0.0757538 |
1
|
410
|
|
GO:0030528
|
transcription regulator activity
|
0.0765699 |
0.0765699 |
1
|
432
|
|
GO:0008347
|
glial cell migration
|
0.0869565 |
0.0869565 |
1
|
24
|
|
GO:0040011
|
locomotion
|
0.0891086 |
0.0891086 |
1
|
484
|
|
GO:0042063
|
gliogenesis
|
0.0901899 |
0.0901899 |
1
|
57
|
|
GO:0010942
|
positive regulation of cell death
|
0.0996310 |
0.0996310 |
1
|
54
|
|
GO:0080134
|
regulation of response to stress
|
0.117925 |
0.117925 |
1
|
75
|
|
GO:0023014
|
signal transmission via phosphorylation event
|
0.126316 |
0.126316 |
1
|
96
|
|
GO:0035466
|
regulation of signaling pathway
|
0.126661 |
0.126661 |
1
|
324
|
|
GO:0010627
|
regulation of intracellular protein kinase cascade
|
0.132075 |
0.132075 |
1
|
49
|
|
GO:0023046
|
signaling process
|
0.138061 |
0.138061 |
1
|
760
|
|
GO:0051674
|
localization of cell
|
0.138713 |
0.138713 |
1
|
263
|
|
GO:0001882
|
nucleoside binding
|
0.138982 |
0.138982 |
1
|
784
|
|
GO:0044427
|
chromosomal part
|
0.156236 |
0.156236 |
1
|
342
|
|
GO:0000775
|
chromosome, centromeric region
|
0.163743 |
0.163743 |
1
|
56
|
|
GO:0031098
|
stress-activated protein kinase signaling cascade
|
0.170886 |
0.170886 |
1
|
54
|
|
GO:0048869
|
cellular developmental process
|
0.176780 |
0.176780 |
1
|
1276
|
|
GO:0032989
|
cellular component morphogenesis
|
0.193108 |
0.193108 |
1
|
566
|
|
GO:0000166
|
nucleotide binding
|
0.208828 |
0.208828 |
1
|
1178
|
|
GO:0004386
|
helicase activity
|
0.210708 |
0.210708 |
1
|
122
|
|
GO:0043068
|
positive regulation of programmed cell death
|
0.225000 |
0.225000 |
1
|
54
|
|
GO:0043065
|
positive regulation of apoptosis
|
0.231250 |
0.231250 |
1
|
37
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.237386 |
0.237386 |
1
|
1562
|
|
GO:0023052
|
signaling
|
0.240473 |
0.240473 |
1
|
1364
|
|
GO:0009605
|
response to external stimulus
|
0.242248 |
0.242248 |
1
|
375
|
|
GO:0007165
|
signal transduction
|
0.254057 |
0.254057 |
1
|
548
|
|
GO:0050896
|
response to stimulus
|
0.270384 |
0.270384 |
1
|
1548
|
|
GO:0005700
|
polytene chromosome
|
0.276650 |
0.276650 |
1
|
109
|
|
GO:0006935
|
chemotaxis
|
0.287630 |
0.287630 |
1
|
193
|
|
GO:0016817
|
hydrolase activity, acting on acid anhydrides
|
0.301217 |
0.301217 |
1
|
594
|
|
GO:0007243
|
intracellular protein kinase cascade
|
0.304762 |
0.304762 |
1
|
96
|
|
GO:0050794
|
regulation of cellular process
|
0.306371 |
0.306371 |
1
|
2034
|
|
GO:0009966
|
regulation of signal transduction
|
0.312044 |
0.312044 |
1
|
171
|
|
GO:0005694
|
chromosome
|
0.321108 |
0.321108 |
1
|
394
|
|
GO:0051179
|
localization
|
0.325313 |
0.325313 |
1
|
1896
|
|
GO:0003678
|
DNA helicase activity
|
0.327869 |
0.327869 |
1
|
40
|
|
GO:0003676
|
nucleic acid binding
|
0.328842 |
0.328842 |
1
|
1855
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.346904 |
0.346904 |
1
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.347592 |
0.347592 |
1
|
1227
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.357722 |
0.357722 |
1
|
2108
|
|
GO:0051276
|
chromosome organization
|
0.358714 |
0.358714 |
1
|
424
|
|
GO:0016251
|
general RNA polymerase II transcription factor activity
|
0.359551 |
0.359551 |
1
|
96
|
|
GO:0000904
|
cell morphogenesis involved in differentiation
|
0.376731 |
0.376731 |
1
|
408
|
|
GO:0050789
|
regulation of biological process
|
0.378390 |
0.378390 |
1
|
2246
|
|
GO:0006350
|
transcription
|
0.387810 |
0.387810 |
1
|
859
|
|
GO:0042330
|
taxis
|
0.389803 |
0.389803 |
1
|
237
|
|
GO:0006950
|
response to stress
|
0.390827 |
0.390827 |
1
|
605
|
|
GO:0033554
|
cellular response to stress
|
0.397122 |
0.397122 |
1
|
276
|
|
GO:0009058
|
biosynthetic process
|
0.402387 |
0.402387 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.402965 |
0.402965 |
1
|
2093
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.405649 |
0.405649 |
1
|
1623
|
|
GO:0042221
|
response to chemical stimulus
|
0.408269 |
0.408269 |
1
|
632
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.415756 |
0.415756 |
1
|
1789
|
|
GO:0065007
|
biological regulation
|
0.422440 |
0.422440 |
1
|
2548
|
|
GO:0023034
|
intracellular signaling pathway
|
0.430962 |
0.430962 |
1
|
412
|
|
GO:0044422
|
organelle part
|
0.432705 |
0.432705 |
1
|
2176
|
|
GO:0048666
|
neuron development
|
0.433702 |
0.433702 |
1
|
471
|
|
GO:0032502
|
developmental process
|
0.435255 |
0.435255 |
1
|
2638
|
|
GO:0043408
|
regulation of MAPKKK cascade
|
0.435294 |
0.435294 |
1
|
37
|
|
GO:0030182
|
neuron differentiation
|
0.442292 |
0.442292 |
1
|
548
|
|
GO:0003677
|
DNA binding
|
0.444205 |
0.444205 |
1
|
824
|
|
GO:0022008
|
neurogenesis
|
0.452398 |
0.452398 |
1
|
632
|
|
GO:0048468
|
cell development
|
0.457419 |
0.457419 |
1
|
1042
|
|
GO:0070302
|
regulation of stress-activated protein kinase signaling cascade
|
0.458333 |
0.458333 |
1
|
33
|
|
GO:0005575
|
cellular_component
|
0.478421 |
0.478421 |
2
|
8817
|
|
GO:0016787
|
hydrolase activity
|
0.480273 |
0.480273 |
1
|
1972
|
|
GO:0044237
|
cellular metabolic process
|
0.490989 |
0.490989 |
1
|
3814
|
|
GO:0044446
|
intracellular organelle part
|
0.493044 |
0.493044 |
1
|
2162
|
|
GO:0005623
|
cell
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.496927 |
0.496927 |
1
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.498346 |
0.498346 |
1
|
1959
|
|
GO:0035556
|
intracellular signal transduction
|
0.499208 |
0.499208 |
1
|
315
|
|
GO:0007399
|
nervous system development
|
0.500000 |
0.500000 |
1
|
848
|
|
GO:0043067
|
regulation of programmed cell death
|
0.509960 |
0.509960 |
1
|
128
|
|
GO:0048870
|
cell motility
|
0.512097 |
0.512097 |
1
|
254
|
|
GO:0044249
|
cellular biosynthetic process
|
0.526807 |
0.526807 |
1
|
2034
|
|
GO:0032501
|
multicellular organismal process
|
0.527040 |
0.527040 |
1
|
3315
|
|
GO:0010467
|
gene expression
|
0.531494 |
0.531494 |
1
|
1907
|
|
GO:0006917
|
induction of apoptosis
|
0.565217 |
0.565217 |
1
|
26
|
|
GO:0016020
|
membrane
|
0.567775 |
0.567775 |
1
|
2122
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0046328
|
regulation of JNK cascade
|
0.589286 |
0.589286 |
1
|
33
|
|
GO:0003824
|
catalytic activity
|
0.604523 |
0.604523 |
1
|
4106
|
|
GO:0003702
|
RNA polymerase II transcription factor activity
|
0.618056 |
0.618056 |
1
|
267
|
|
GO:0006915
|
apoptosis
|
0.633333 |
0.633333 |
1
|
152
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.639129 |
0.639129 |
1
|
2878
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0043226
|
organelle
|
0.641414 |
0.641414 |
1
|
3537
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0012502
|
induction of programmed cell death
|
0.666667 |
0.666667 |
1
|
36
|
|
GO:0042981
|
regulation of apoptosis
|
0.674699 |
0.674699 |
1
|
112
|
|
GO:0007409
|
axonogenesis
|
0.679389 |
0.679389 |
1
|
267
|
|
GO:0007411
|
axon guidance
|
0.683824 |
0.683824 |
1
|
186
|
|
GO:0043170
|
macromolecule metabolic process
|
0.690797 |
0.690797 |
1
|
3588
|
|
GO:0008150
|
biological_process
|
0.693584 |
0.693584 |
2
|
10616
|
|
GO:0031175
|
neuron projection development
|
0.696270 |
0.696270 |
1
|
392
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0007254
|
JNK cascade
|
0.702703 |
0.702703 |
1
|
52
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.724105 |
0.724105 |
1
|
1517
|
|
GO:0008152
|
metabolic process
|
0.739170 |
0.739170 |
1
|
5194
|
|
GO:0032990
|
cell part morphogenesis
|
0.745583 |
0.745583 |
1
|
422
|
|
GO:0032553
|
ribonucleotide binding
|
0.746180 |
0.746180 |
1
|
879
|
|
GO:0003674
|
molecular_function
|
0.753361 |
0.753361 |
2
|
11064
|
|
GO:0032559
|
adenyl ribonucleotide binding
|
0.757705 |
0.757705 |
1
|
713
|
|
GO:0005488
|
binding
|
0.759777 |
0.759777 |
1
|
5641
|
|
GO:0000165
|
MAPKKK cascade
|
0.760417 |
0.760417 |
1
|
73
|
|
GO:0006996
|
organelle organization
|
0.779169 |
0.779169 |
1
|
1182
|
|
GO:0048667
|
cell morphogenesis involved in neuron differentiation
|
0.795501 |
0.795501 |
1
|
389
|
|
GO:0017076
|
purine nucleotide binding
|
0.803056 |
0.803056 |
1
|
946
|
|
GO:0043229
|
intracellular organelle
|
0.804100 |
0.804100 |
1
|
3530
|
|
GO:0048858
|
cell projection morphogenesis
|
0.806883 |
0.806883 |
1
|
422
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0030554
|
adenyl nucleotide binding
|
0.819239 |
0.819239 |
1
|
775
|
|
GO:0044238
|
primary metabolic process
|
0.819985 |
0.819985 |
1
|
4259
|
|
GO:0000902
|
cell morphogenesis
|
0.825088 |
0.825088 |
1
|
467
|
|
GO:0009987
|
cellular process
|
0.854049 |
0.854049 |
1
|
6560
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0048812
|
neuron projection morphogenesis
|
0.910798 |
0.910798 |
1
|
388
|
|
GO:0044424
|
intracellular part
|
0.914575 |
0.914575 |
1
|
4384
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.929175 |
0.929175 |
1
|
879
|
|
GO:0012501
|
programmed cell death
|
0.941176 |
0.941176 |
1
|
240
|
|
GO:0005622
|
intracellular
|
0.944411 |
0.944411 |
1
|
4734
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0016477
|
cell migration
|
0.964567 |
0.964567 |
1
|
245
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0017111
|
nucleoside-triphosphatase activity
|
0.983022 |
0.983022 |
1
|
579
|
|
GO:0016462
|
pyrophosphatase activity
|
0.991582 |
0.991582 |
1
|
589
|
|
GO:0001883
|
purine nucleoside binding
|
0.992347 |
0.992347 |
1
|
778
|
|
GO:0005524
|
ATP binding
|
0.998597 |
0.998597 |
1
|
712
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
2
|
12747
|
|
GO:0016818
|
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
|
1.00000 |
1.00000 |
1
|
594
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|