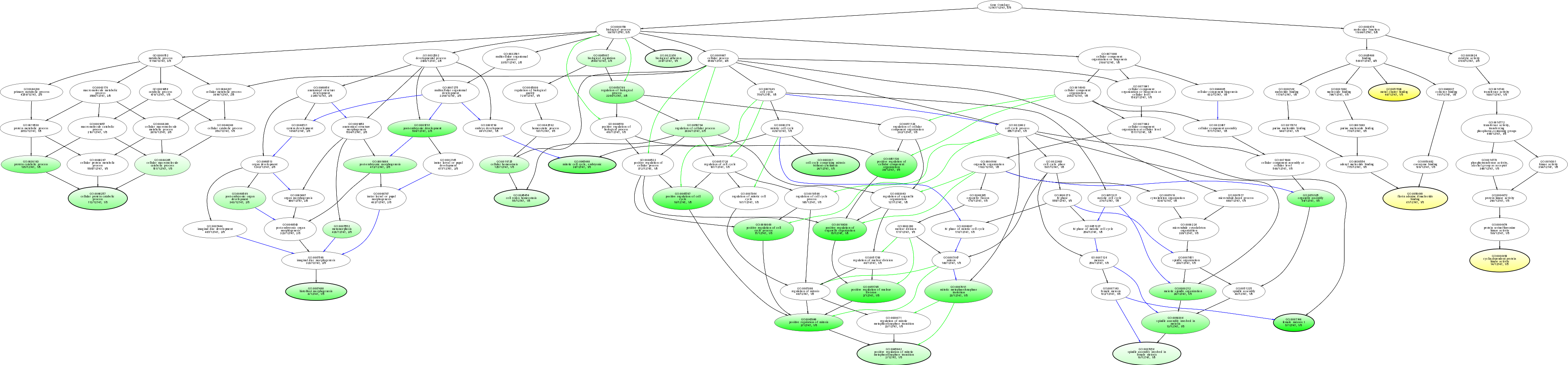
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0007144
|
female meiosis I
|
0.00763359 |
0.00763359 |
1
|
5
|
|
GO:0010638
|
positive regulation of organelle organization
|
0.0103448 |
0.0103448 |
1
|
15
|
|
GO:0045840
|
positive regulation of mitosis
|
0.0106383 |
0.0106383 |
1
|
2
|
|
GO:0051785
|
positive regulation of nuclear division
|
0.0109290 |
0.0109290 |
1
|
2
|
|
GO:0051130
|
positive regulation of cellular component organization
|
0.0150642 |
0.0150642 |
1
|
34
|
|
GO:0045787
|
positive regulation of cell cycle
|
0.0171103 |
0.0171103 |
1
|
18
|
|
GO:0090068
|
positive regulation of cell cycle process
|
0.0175620 |
0.0175620 |
1
|
17
|
|
GO:0051540
|
metal cluster binding
|
0.0322300 |
0.0322300 |
1
|
46
|
|
GO:0007091
|
mitotic metaphase/anaphase transition
|
0.0351145 |
0.0351145 |
1
|
23
|
|
GO:0009791
|
post-embryonic development
|
0.0358662 |
0.0358662 |
2
|
500
|
|
GO:0045448
|
mitotic cell cycle, embryonic
|
0.0363636 |
0.0363636 |
1
|
36
|
|
GO:0070925
|
organelle assembly
|
0.0383299 |
0.0383299 |
1
|
56
|
|
GO:0000212
|
meiotic spindle organization
|
0.0426829 |
0.0426829 |
1
|
28
|
|
GO:0090306
|
spindle assembly involved in meiosis
|
0.0454545 |
0.0454545 |
1
|
13
|
|
GO:0007488
|
histoblast morphogenesis
|
0.0489972 |
0.0489972 |
1
|
8
|
|
GO:0033301
|
cell cycle comprising mitosis without cytokinesis
|
0.0559441 |
0.0559441 |
1
|
24
|
|
GO:0030163
|
protein catabolic process
|
0.0636192 |
0.0636192 |
1
|
135
|
|
GO:0050789
|
regulation of biological process
|
0.0671442 |
0.0671442 |
3
|
2246
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.0671911 |
0.0671911 |
2
|
412
|
|
GO:0004693
|
cyclin-dependent protein kinase activity
|
0.0703518 |
0.0703518 |
1
|
14
|
|
GO:0050660
|
flavin adenine dinucleotide binding
|
0.0715123 |
0.0715123 |
1
|
61
|
|
GO:0044257
|
cellular protein catabolic process
|
0.0719332 |
0.0719332 |
1
|
112
|
|
GO:0019725
|
cellular homeostasis
|
0.0740521 |
0.0740521 |
1
|
125
|
|
GO:0007552
|
metamorphosis
|
0.0748335 |
0.0748335 |
2
|
420
|
|
GO:0048569
|
post-embryonic organ development
|
0.0761074 |
0.0761074 |
2
|
343
|
|
GO:0045454
|
cell redox homeostasis
|
0.0770675 |
0.0770675 |
1
|
55
|
|
GO:0045842
|
positive regulation of mitotic metaphase/anaphase transition
|
0.0869565 |
0.0869565 |
1
|
2
|
|
GO:0050794
|
regulation of cellular process
|
0.0885337 |
0.0885337 |
3
|
2034
|
|
GO:0065007
|
biological regulation
|
0.0932173 |
0.0932173 |
3
|
2548
|
|
GO:0022610
|
biological adhesion
|
0.0933472 |
0.0933472 |
1
|
206
|
|
GO:0044265
|
cellular macromolecule catabolic process
|
0.0950019 |
0.0950019 |
1
|
151
|
|
GO:0007056
|
spindle assembly involved in female meiosis
|
0.0952381 |
0.0952381 |
1
|
10
|
|
GO:0009057
|
macromolecule catabolic process
|
0.107103 |
0.107103 |
1
|
210
|
|
GO:0007155
|
cell adhesion
|
0.111846 |
0.111846 |
1
|
192
|
|
GO:0048037
|
cofactor binding
|
0.128745 |
0.128745 |
1
|
191
|
|
GO:0033043
|
regulation of organelle organization
|
0.130571 |
0.130571 |
1
|
127
|
|
GO:0009887
|
organ morphogenesis
|
0.140035 |
0.140035 |
2
|
684
|
|
GO:0007444
|
imaginal disc development
|
0.141192 |
0.141192 |
2
|
467
|
|
GO:0016563
|
transcription activator activity
|
0.141204 |
0.141204 |
1
|
61
|
|
GO:0010564
|
regulation of cell cycle process
|
0.148725 |
0.148725 |
1
|
105
|
|
GO:0030234
|
enzyme regulator activity
|
0.148898 |
0.148898 |
1
|
351
|
|
GO:0048285
|
organelle fission
|
0.148900 |
0.148900 |
1
|
176
|
|
GO:0007057
|
spindle assembly involved in female meiosis I
|
0.153846 |
0.153846 |
1
|
2
|
|
GO:0051225
|
spindle assembly
|
0.159363 |
0.159363 |
1
|
40
|
|
GO:0043161
|
proteasomal ubiquitin-dependent protein catabolic process
|
0.161616 |
0.161616 |
1
|
16
|
|
GO:0019207
|
kinase regulator activity
|
0.162393 |
0.162393 |
1
|
57
|
|
GO:0051603
|
proteolysis involved in cellular protein catabolic process
|
0.162791 |
0.162791 |
1
|
112
|
|
GO:0048563
|
post-embryonic organ morphogenesis
|
0.172051 |
0.172051 |
2
|
323
|
|
GO:0044248
|
cellular catabolic process
|
0.172381 |
0.172381 |
1
|
350
|
|
GO:0051537
|
2 iron, 2 sulfur cluster binding
|
0.173913 |
0.173913 |
1
|
8
|
|
GO:0010498
|
proteasomal protein catabolic process
|
0.178571 |
0.178571 |
1
|
20
|
|
GO:0048518
|
positive regulation of biological process
|
0.178785 |
0.178785 |
1
|
410
|
|
GO:0030528
|
transcription regulator activity
|
0.180596 |
0.180596 |
1
|
432
|
|
GO:0051783
|
regulation of nuclear division
|
0.185484 |
0.185484 |
1
|
46
|
|
GO:0007088
|
regulation of mitosis
|
0.187755 |
0.187755 |
1
|
46
|
|
GO:0051716
|
cellular response to stimulus
|
0.195075 |
0.195075 |
1
|
374
|
|
GO:0005488
|
binding
|
0.200001 |
0.200001 |
4
|
5641
|
|
GO:0048522
|
positive regulation of cellular process
|
0.207979 |
0.207979 |
1
|
372
|
|
GO:0051726
|
regulation of cell cycle
|
0.209192 |
0.209192 |
1
|
187
|
|
GO:0048736
|
appendage development
|
0.209532 |
0.209532 |
1
|
286
|
|
GO:0007346
|
regulation of mitotic cell cycle
|
0.220165 |
0.220165 |
1
|
107
|
|
GO:0051128
|
regulation of cellular component organization
|
0.225720 |
0.225720 |
1
|
282
|
|
GO:0060089
|
molecular transducer activity
|
0.228389 |
0.228389 |
1
|
559
|
|
GO:0007017
|
microtubule-based process
|
0.246490 |
0.246490 |
1
|
448
|
|
GO:0009056
|
catabolic process
|
0.251979 |
0.251979 |
1
|
479
|
|
GO:0042592
|
homeostatic process
|
0.256516 |
0.256516 |
1
|
187
|
|
GO:0055114
|
oxidation-reduction process
|
0.269455 |
0.269455 |
1
|
516
|
|
GO:0000087
|
M phase of mitotic cell cycle
|
0.269525 |
0.269525 |
1
|
176
|
|
GO:0006793
|
phosphorus metabolic process
|
0.270339 |
0.270339 |
1
|
556
|
|
GO:0022607
|
cellular component assembly
|
0.275418 |
0.275418 |
1
|
577
|
|
GO:0007067
|
mitosis
|
0.281356 |
0.281356 |
1
|
166
|
|
GO:0016491
|
oxidoreductase activity
|
0.286653 |
0.286653 |
1
|
638
|
|
GO:0032774
|
RNA biosynthetic process
|
0.290017 |
0.290017 |
1
|
703
|
|
GO:0044085
|
cellular component biogenesis
|
0.299810 |
0.299810 |
1
|
632
|
|
GO:0048513
|
organ development
|
0.300572 |
0.300572 |
2
|
1242
|
|
GO:0023046
|
signaling process
|
0.310289 |
0.310289 |
1
|
760
|
|
GO:0016538
|
cyclin-dependent protein kinase regulator activity
|
0.327273 |
0.327273 |
1
|
18
|
|
GO:0007560
|
imaginal disc morphogenesis
|
0.331061 |
0.331061 |
2
|
323
|
|
GO:0035107
|
appendage morphogenesis
|
0.334837 |
0.334837 |
1
|
283
|
|
GO:0006508
|
proteolysis
|
0.335119 |
0.335119 |
1
|
688
|
|
GO:0006468
|
protein phosphorylation
|
0.335758 |
0.335758 |
1
|
277
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.338051 |
0.338051 |
2
|
1534
|
|
GO:0022402
|
cell cycle process
|
0.343522 |
0.343522 |
1
|
655
|
|
GO:0051321
|
meiotic cell cycle
|
0.357143 |
0.357143 |
1
|
270
|
|
GO:0005515
|
protein binding
|
0.358840 |
0.358840 |
2
|
1726
|
|
GO:0043412
|
macromolecule modification
|
0.362896 |
0.362896 |
1
|
724
|
|
GO:0032502
|
developmental process
|
0.364008 |
0.364008 |
2
|
2638
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.366791 |
0.366791 |
1
|
888
|
|
GO:0007051
|
spindle organization
|
0.368421 |
0.368421 |
1
|
238
|
|
GO:0009987
|
cellular process
|
0.368587 |
0.368587 |
4
|
6560
|
|
GO:0071844
|
cellular component assembly at cellular level
|
0.373193 |
0.373193 |
1
|
568
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.381695 |
0.381695 |
1
|
905
|
|
GO:0007049
|
cell cycle
|
0.387308 |
0.387308 |
1
|
756
|
|
GO:0006350
|
transcription
|
0.387810 |
0.387810 |
1
|
859
|
|
GO:0006950
|
response to stress
|
0.390827 |
0.390827 |
1
|
605
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.396252 |
0.396252 |
1
|
888
|
|
GO:0033554
|
cellular response to stress
|
0.397122 |
0.397122 |
1
|
276
|
|
GO:0008150
|
biological_process
|
0.400586 |
0.400586 |
5
|
10616
|
|
GO:0007143
|
female meiosis
|
0.401575 |
0.401575 |
1
|
102
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.408435 |
0.408435 |
2
|
2878
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.418200 |
0.418200 |
1
|
1039
|
|
GO:0009790
|
embryo development
|
0.427001 |
0.427001 |
1
|
641
|
|
GO:0048731
|
system development
|
0.432376 |
0.432376 |
2
|
1696
|
|
GO:0007126
|
meiosis
|
0.432709 |
0.432709 |
1
|
254
|
|
GO:0008054
|
cyclin catabolic process
|
0.437500 |
0.437500 |
1
|
7
|
|
GO:0016740
|
transferase activity
|
0.438824 |
0.438824 |
1
|
1030
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.440308 |
0.440308 |
1
|
686
|
|
GO:0007275
|
multicellular organismal development
|
0.442066 |
0.442066 |
2
|
2296
|
|
GO:0006464
|
protein modification process
|
0.443005 |
0.443005 |
1
|
684
|
|
GO:0001882
|
nucleoside binding
|
0.450493 |
0.450493 |
1
|
784
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.451726 |
0.451726 |
1
|
903
|
|
GO:0007010
|
cytoskeleton organization
|
0.453469 |
0.453469 |
1
|
536
|
|
GO:0051327
|
M phase of meiotic cell cycle
|
0.456014 |
0.456014 |
1
|
254
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.456989 |
0.456989 |
1
|
850
|
|
GO:0008152
|
metabolic process
|
0.479868 |
0.479868 |
3
|
5194
|
|
GO:0016772
|
transferase activity, transferring phosphorus-containing groups
|
0.480583 |
0.480583 |
1
|
495
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.486014 |
0.486014 |
1
|
1056
|
|
GO:0010468
|
regulation of gene expression
|
0.489930 |
0.489930 |
1
|
973
|
|
GO:0003674
|
molecular_function
|
0.492571 |
0.492571 |
5
|
11064
|
|
GO:0032501
|
multicellular organismal process
|
0.496887 |
0.496887 |
2
|
3315
|
|
GO:0023052
|
signaling
|
0.497295 |
0.497295 |
1
|
1364
|
|
GO:0030071
|
regulation of mitotic metaphase/anaphase transition
|
0.500000 |
0.500000 |
1
|
23
|
|
GO:0044424
|
intracellular part
|
0.500760 |
0.500760 |
2
|
4384
|
|
GO:0019222
|
regulation of metabolic process
|
0.507779 |
0.507779 |
1
|
1242
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.513595 |
0.513595 |
1
|
850
|
|
GO:0000226
|
microtubule cytoskeleton organization
|
0.529687 |
0.529687 |
1
|
339
|
|
GO:0050896
|
response to stimulus
|
0.545344 |
0.545344 |
1
|
1548
|
|
GO:0000278
|
mitotic cell cycle
|
0.567460 |
0.567460 |
1
|
429
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.569226 |
0.569226 |
1
|
1110
|
|
GO:0035186
|
syncytial blastoderm mitotic cell cycle
|
0.583333 |
0.583333 |
1
|
21
|
|
GO:0005622
|
intracellular
|
0.583919 |
0.583919 |
2
|
4734
|
|
GO:0007165
|
signal transduction
|
0.585130 |
0.585130 |
1
|
548
|
|
GO:0000166
|
nucleotide binding
|
0.608293 |
0.608293 |
1
|
1178
|
|
GO:0003824
|
catalytic activity
|
0.611425 |
0.611425 |
2
|
4106
|
|
GO:0003702
|
RNA polymerase II transcription factor activity
|
0.618056 |
0.618056 |
1
|
267
|
|
GO:0065008
|
regulation of biological quality
|
0.636341 |
0.636341 |
1
|
729
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.646807 |
0.646807 |
1
|
1623
|
|
GO:0016070
|
RNA metabolic process
|
0.656488 |
0.656488 |
1
|
1191
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.658716 |
0.658716 |
1
|
1789
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.661861 |
0.661861 |
1
|
1562
|
|
GO:0043632
|
modification-dependent macromolecule catabolic process
|
0.662252 |
0.662252 |
1
|
100
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.669454 |
0.669454 |
1
|
2108
|
|
GO:0044237
|
cellular metabolic process
|
0.673909 |
0.673909 |
2
|
3814
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.684327 |
0.684327 |
1
|
620
|
|
GO:0007423
|
sensory organ development
|
0.684358 |
0.684358 |
1
|
544
|
|
GO:0004674
|
protein serine/threonine kinase activity
|
0.686207 |
0.686207 |
1
|
199
|
|
GO:0044267
|
cellular protein metabolic process
|
0.686538 |
0.686538 |
1
|
1505
|
|
GO:0045449
|
regulation of transcription
|
0.687166 |
0.687166 |
1
|
771
|
|
GO:0006974
|
response to DNA damage stimulus
|
0.706522 |
0.706522 |
1
|
195
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.709978 |
0.709978 |
2
|
402
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.724105 |
0.724105 |
1
|
1517
|
|
GO:0019538
|
protein metabolic process
|
0.724907 |
0.724907 |
1
|
2053
|
|
GO:0050662
|
coenzyme binding
|
0.727749 |
0.727749 |
1
|
139
|
|
GO:0004672
|
protein kinase activity
|
0.728643 |
0.728643 |
1
|
290
|
|
GO:0016773
|
phosphotransferase activity, alcohol group as acceptor
|
0.735354 |
0.735354 |
1
|
364
|
|
GO:0048856
|
anatomical structure development
|
0.737157 |
0.737157 |
2
|
2265
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.746994 |
0.746994 |
1
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.748407 |
0.748407 |
1
|
1959
|
|
GO:0016310
|
phosphorylation
|
0.758993 |
0.758993 |
1
|
422
|
|
GO:0043170
|
macromolecule metabolic process
|
0.772351 |
0.772351 |
2
|
3588
|
|
GO:0044249
|
cellular biosynthetic process
|
0.776153 |
0.776153 |
1
|
2034
|
|
GO:0006996
|
organelle organization
|
0.779169 |
0.779169 |
1
|
1182
|
|
GO:0010467
|
gene expression
|
0.780571 |
0.780571 |
1
|
1907
|
|
GO:0043226
|
organelle
|
0.785296 |
0.785296 |
1
|
3537
|
|
GO:0009058
|
biosynthetic process
|
0.786651 |
0.786651 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.787269 |
0.787269 |
1
|
2093
|
|
GO:0005623
|
cell
|
0.787322 |
0.787322 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.787322 |
0.787322 |
2
|
6195
|
|
GO:0005737
|
cytoplasm
|
0.790684 |
0.790684 |
1
|
2378
|
|
GO:0016301
|
kinase activity
|
0.795960 |
0.795960 |
1
|
394
|
|
GO:0017076
|
purine nucleotide binding
|
0.803056 |
0.803056 |
1
|
946
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.814170 |
0.814170 |
1
|
701
|
|
GO:0030554
|
adenyl nucleotide binding
|
0.819239 |
0.819239 |
1
|
775
|
|
GO:0005575
|
cellular_component
|
0.825797 |
0.825797 |
3
|
8817
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0035120
|
post-embryonic appendage morphogenesis
|
0.869576 |
0.869576 |
1
|
268
|
|
GO:0019941
|
modification-dependent protein catabolic process
|
0.876106 |
0.876106 |
1
|
99
|
|
GO:0007472
|
wing disc morphogenesis
|
0.888890 |
0.888890 |
1
|
255
|
|
GO:0022403
|
cell cycle phase
|
0.896183 |
0.896183 |
1
|
587
|
|
GO:0035220
|
wing disc development
|
0.898668 |
0.898668 |
1
|
318
|
|
GO:0007476
|
imaginal disc-derived wing morphogenesis
|
0.903915 |
0.903915 |
1
|
254
|
|
GO:0002165
|
instar larval or pupal development
|
0.910028 |
0.910028 |
2
|
477
|
|
GO:0044238
|
primary metabolic process
|
0.914505 |
0.914505 |
2
|
4259
|
|
GO:0000279
|
M phase
|
0.945486 |
0.945486 |
1
|
555
|
|
GO:0043229
|
intracellular organelle
|
0.961659 |
0.961659 |
1
|
3530
|
|
GO:0019887
|
protein kinase regulator activity
|
0.964912 |
0.964912 |
1
|
55
|
|
GO:0000280
|
nuclear division
|
0.971591 |
0.971591 |
1
|
171
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0035114
|
imaginal disc-derived appendage morphogenesis
|
0.975524 |
0.975524 |
1
|
279
|
|
GO:0006511
|
ubiquitin-dependent protein catabolic process
|
0.979798 |
0.979798 |
1
|
97
|
|
GO:0048737
|
imaginal disc-derived appendage development
|
0.986014 |
0.986014 |
1
|
282
|
|
GO:0001883
|
purine nucleoside binding
|
0.992347 |
0.992347 |
1
|
778
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
5
|
12747
|
|
GO:0051536
|
iron-sulfur cluster binding
|
1.00000 |
1.00000 |
1
|
46
|
|
GO:0004871
|
signal transducer activity
|
1.00000 |
1.00000 |
1
|
559
|
|
GO:0006796
|
phosphate metabolic process
|
1.00000 |
1.00000 |
1
|
556
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|