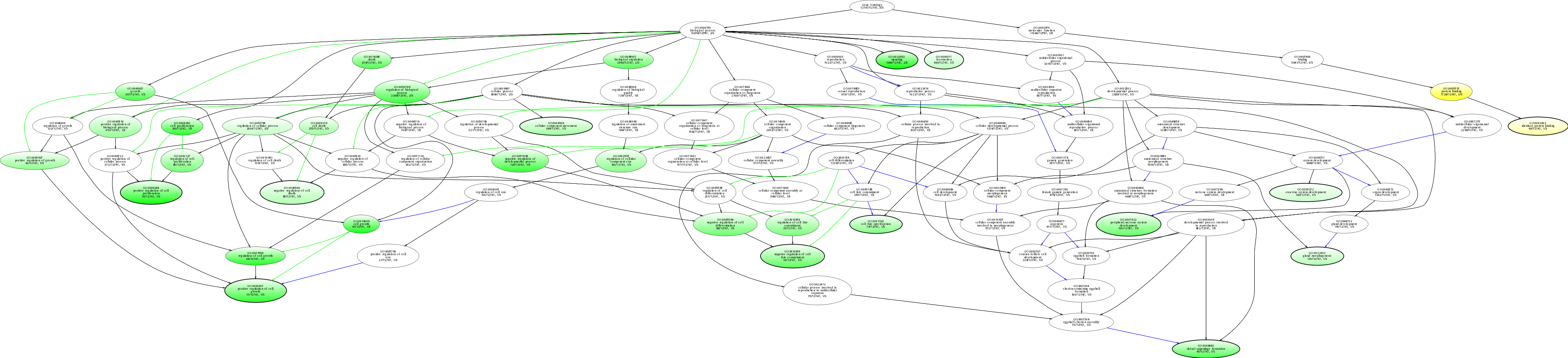
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0023052
|
signaling
|
0.0164979 |
0.0164979 |
2
|
1364
|
|
GO:0008284
|
positive regulation of cell proliferation
|
0.0269750 |
0.0269750 |
1
|
14
|
|
GO:0016049
|
cell growth
|
0.0274662 |
0.0274662 |
1
|
91
|
|
GO:0005515
|
protein binding
|
0.0286108 |
0.0286108 |
3
|
1726
|
|
GO:0001558
|
regulation of cell growth
|
0.0363519 |
0.0363519 |
1
|
38
|
|
GO:0008283
|
cell proliferation
|
0.0373258 |
0.0373258 |
1
|
200
|
|
GO:0030307
|
positive regulation of cell growth
|
0.0376940 |
0.0376940 |
1
|
17
|
|
GO:0051093
|
negative regulation of developmental process
|
0.0423208 |
0.0423208 |
1
|
124
|
|
GO:0050789
|
regulation of biological process
|
0.0447451 |
0.0447451 |
2
|
2246
|
|
GO:0040007
|
growth
|
0.0459943 |
0.0459943 |
1
|
247
|
|
GO:0016265
|
death
|
0.0482013 |
0.0482013 |
1
|
259
|
|
GO:0046843
|
dorsal appendage formation
|
0.0500626 |
0.0500626 |
1
|
40
|
|
GO:0065007
|
biological regulation
|
0.0575901 |
0.0575901 |
2
|
2548
|
|
GO:0010454
|
negative regulation of cell fate commitment
|
0.0584416 |
0.0584416 |
1
|
18
|
|
GO:0007422
|
peripheral nervous system development
|
0.0589623 |
0.0589623 |
1
|
100
|
|
GO:0045596
|
negative regulation of cell differentiation
|
0.0604154 |
0.0604154 |
1
|
96
|
|
GO:0042127
|
regulation of cell proliferation
|
0.0612481 |
0.0612481 |
1
|
65
|
|
GO:0010453
|
regulation of cell fate commitment
|
0.0612745 |
0.0612745 |
1
|
25
|
|
GO:0001708
|
cell fate specification
|
0.0619122 |
0.0619122 |
1
|
79
|
|
GO:0032535
|
regulation of cellular component size
|
0.0631681 |
0.0631681 |
1
|
130
|
|
GO:0045927
|
positive regulation of growth
|
0.0684932 |
0.0684932 |
1
|
40
|
|
GO:0048518
|
positive regulation of biological process
|
0.0757538 |
0.0757538 |
1
|
410
|
|
GO:0008219
|
cell death
|
0.0762386 |
0.0762386 |
1
|
255
|
|
GO:0006928
|
cellular component movement
|
0.0803367 |
0.0803367 |
1
|
269
|
|
GO:0042802
|
identical protein binding
|
0.0811770 |
0.0811770 |
1
|
48
|
|
GO:0060548
|
negative regulation of cell death
|
0.0821745 |
0.0821745 |
1
|
65
|
|
GO:0022612
|
gland morphogenesis
|
0.0873786 |
0.0873786 |
1
|
135
|
|
GO:0040011
|
locomotion
|
0.0891086 |
0.0891086 |
1
|
484
|
|
GO:0050794
|
regulation of cellular process
|
0.0938314 |
0.0938314 |
2
|
2034
|
|
GO:0035272
|
exocrine system development
|
0.0955189 |
0.0955189 |
1
|
162
|
|
GO:0001071
|
nucleic acid binding transcription factor activity
|
0.103335 |
0.103335 |
1
|
395
|
|
GO:0001709
|
cell fate determination
|
0.103448 |
0.103448 |
1
|
132
|
|
GO:0030703
|
eggshell formation
|
0.104733 |
0.104733 |
1
|
104
|
|
GO:0048522
|
positive regulation of cellular process
|
0.110028 |
0.110028 |
1
|
372
|
|
GO:0007306
|
eggshell chorion assembly
|
0.110942 |
0.110942 |
1
|
73
|
|
GO:0040008
|
regulation of growth
|
0.111607 |
0.111607 |
1
|
132
|
|
GO:0030528
|
transcription regulator activity
|
0.112632 |
0.112632 |
1
|
432
|
|
GO:0008593
|
regulation of Notch signaling pathway
|
0.112676 |
0.112676 |
1
|
40
|
|
GO:0019899
|
enzyme binding
|
0.113660 |
0.113660 |
1
|
68
|
|
GO:0007219
|
Notch signaling pathway
|
0.118590 |
0.118590 |
1
|
74
|
|
GO:0022412
|
cellular process involved in reproduction in multicellular organism
|
0.122349 |
0.122349 |
1
|
75
|
|
GO:0045746
|
negative regulation of Notch signaling pathway
|
0.124260 |
0.124260 |
1
|
21
|
|
GO:0007297
|
ovarian follicle cell migration
|
0.126918 |
0.126918 |
1
|
91
|
|
GO:0010941
|
regulation of cell death
|
0.127443 |
0.127443 |
1
|
139
|
|
GO:0048519
|
negative regulation of biological process
|
0.128590 |
0.128590 |
1
|
706
|
|
GO:0005488
|
binding
|
0.132501 |
0.132501 |
3
|
5641
|
|
GO:0010927
|
cellular component assembly involved in morphogenesis
|
0.134157 |
0.134157 |
1
|
152
|
|
GO:0007154
|
cell communication
|
0.135620 |
0.135620 |
1
|
461
|
|
GO:0010648
|
negative regulation of cell communication
|
0.136511 |
0.136511 |
1
|
126
|
|
GO:0023046
|
signaling process
|
0.138061 |
0.138061 |
1
|
760
|
|
GO:0051674
|
localization of cell
|
0.138713 |
0.138713 |
1
|
263
|
|
GO:0046983
|
protein dimerization activity
|
0.150059 |
0.150059 |
1
|
91
|
|
GO:0048732
|
gland development
|
0.153784 |
0.153784 |
1
|
191
|
|
GO:0007559
|
histolysis
|
0.156194 |
0.156194 |
1
|
87
|
|
GO:0051128
|
regulation of cellular component organization
|
0.156775 |
0.156775 |
1
|
282
|
|
GO:0050793
|
regulation of developmental process
|
0.161873 |
0.161873 |
1
|
321
|
|
GO:0045595
|
regulation of cell differentiation
|
0.167859 |
0.167859 |
1
|
231
|
|
GO:0030707
|
ovarian follicle cell development
|
0.170695 |
0.170695 |
1
|
226
|
|
GO:0048610
|
cellular process involved in reproduction
|
0.173346 |
0.173346 |
1
|
613
|
|
GO:0035070
|
salivary gland histolysis
|
0.175325 |
0.175325 |
1
|
81
|
|
GO:0048646
|
anatomical structure formation involved in morphogenesis
|
0.176649 |
0.176649 |
1
|
466
|
|
GO:0000003
|
reproduction
|
0.183280 |
0.183280 |
1
|
1022
|
|
GO:0022414
|
reproductive process
|
0.183280 |
0.183280 |
1
|
1022
|
|
GO:0048523
|
negative regulation of cellular process
|
0.183975 |
0.183975 |
1
|
635
|
|
GO:0010646
|
regulation of cell communication
|
0.186653 |
0.186653 |
1
|
214
|
|
GO:0009791
|
post-embryonic development
|
0.189538 |
0.189538 |
1
|
500
|
|
GO:0032989
|
cellular component morphogenesis
|
0.193108 |
0.193108 |
1
|
566
|
|
GO:0045165
|
cell fate commitment
|
0.199843 |
0.199843 |
1
|
255
|
|
GO:0007389
|
pattern specification process
|
0.203563 |
0.203563 |
1
|
537
|
|
GO:0003006
|
developmental process involved in reproduction
|
0.204901 |
0.204901 |
1
|
602
|
|
GO:0045793
|
positive regulation of cell size
|
0.210000 |
0.210000 |
1
|
21
|
|
GO:0009996
|
negative regulation of cell fate specification
|
0.227848 |
0.227848 |
1
|
18
|
|
GO:0043069
|
negative regulation of programmed cell death
|
0.229839 |
0.229839 |
1
|
57
|
|
GO:0035467
|
negative regulation of signaling pathway
|
0.230613 |
0.230613 |
1
|
118
|
|
GO:0090066
|
regulation of anatomical structure size
|
0.231824 |
0.231824 |
1
|
169
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.237215 |
0.237215 |
1
|
1039
|
|
GO:0035466
|
regulation of signaling pathway
|
0.237323 |
0.237323 |
1
|
324
|
|
GO:0009888
|
tissue development
|
0.245916 |
0.245916 |
1
|
557
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.259446 |
0.259446 |
1
|
412
|
|
GO:0008101
|
decapentaplegic receptor signaling pathway
|
0.265625 |
0.265625 |
1
|
17
|
|
GO:0007552
|
metamorphosis
|
0.273794 |
0.273794 |
1
|
420
|
|
GO:0022607
|
cellular component assembly
|
0.275418 |
0.275418 |
1
|
577
|
|
GO:0048609
|
multicellular organismal reproductive process
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0032504
|
multicellular organism reproduction
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.283034 |
0.283034 |
1
|
1056
|
|
GO:0032774
|
RNA biosynthetic process
|
0.290017 |
0.290017 |
1
|
703
|
|
GO:0046530
|
photoreceptor cell differentiation
|
0.295620 |
0.295620 |
1
|
162
|
|
GO:0044085
|
cellular component biogenesis
|
0.299810 |
0.299810 |
1
|
632
|
|
GO:0042659
|
regulation of cell fate specification
|
0.300000 |
0.300000 |
1
|
24
|
|
GO:0007167
|
enzyme linked receptor protein signaling pathway
|
0.315705 |
0.315705 |
1
|
197
|
|
GO:0048102
|
autophagic cell death
|
0.316667 |
0.316667 |
1
|
76
|
|
GO:0048869
|
cellular developmental process
|
0.322329 |
0.322329 |
1
|
1276
|
|
GO:0043066
|
negative regulation of apoptosis
|
0.322785 |
0.322785 |
1
|
51
|
|
GO:0007178
|
transmembrane receptor protein serine/threonine kinase signaling pathway
|
0.324873 |
0.324873 |
1
|
64
|
|
GO:0051179
|
localization
|
0.325313 |
0.325313 |
1
|
1896
|
|
GO:0016271
|
tissue death
|
0.335907 |
0.335907 |
1
|
87
|
|
GO:0001751
|
compound eye photoreceptor cell differentiation
|
0.354497 |
0.354497 |
1
|
134
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.357722 |
0.357722 |
1
|
2108
|
|
GO:0001754
|
eye photoreceptor cell differentiation
|
0.361386 |
0.361386 |
1
|
146
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.366791 |
0.366791 |
1
|
888
|
|
GO:0071844
|
cellular component assembly at cellular level
|
0.373193 |
0.373193 |
1
|
568
|
|
GO:0009887
|
organ morphogenesis
|
0.374384 |
0.374384 |
1
|
684
|
|
GO:0019222
|
regulation of metabolic process
|
0.376562 |
0.376562 |
1
|
1242
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.381695 |
0.381695 |
1
|
905
|
|
GO:0009987
|
cellular process
|
0.381822 |
0.381822 |
2
|
6560
|
|
GO:0006350
|
transcription
|
0.387810 |
0.387810 |
1
|
859
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.396252 |
0.396252 |
1
|
888
|
|
GO:0043226
|
organelle
|
0.401157 |
0.401157 |
1
|
3537
|
|
GO:0009058
|
biosynthetic process
|
0.402387 |
0.402387 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.402965 |
0.402965 |
1
|
2093
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.405649 |
0.405649 |
1
|
1623
|
|
GO:0016070
|
RNA metabolic process
|
0.413829 |
0.413829 |
1
|
1191
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.415756 |
0.415756 |
1
|
1789
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.418447 |
0.418447 |
1
|
1562
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.429576 |
0.429576 |
1
|
1110
|
|
GO:0032502
|
developmental process
|
0.435255 |
0.435255 |
1
|
2638
|
|
GO:0007423
|
sensory organ development
|
0.438003 |
0.438003 |
1
|
544
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.440308 |
0.440308 |
1
|
686
|
|
GO:0051020
|
GTPase binding
|
0.441176 |
0.441176 |
1
|
30
|
|
GO:0030182
|
neuron differentiation
|
0.442292 |
0.442292 |
1
|
548
|
|
GO:0007165
|
signal transduction
|
0.443656 |
0.443656 |
1
|
548
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.451726 |
0.451726 |
1
|
903
|
|
GO:0022008
|
neurogenesis
|
0.452398 |
0.452398 |
1
|
632
|
|
GO:0007304
|
chorion-containing eggshell formation
|
0.453744 |
0.453744 |
1
|
103
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.456989 |
0.456989 |
1
|
850
|
|
GO:0048468
|
cell development
|
0.457419 |
0.457419 |
1
|
1042
|
|
GO:0010468
|
regulation of gene expression
|
0.489930 |
0.489930 |
1
|
973
|
|
GO:0065008
|
regulation of biological quality
|
0.490437 |
0.490437 |
1
|
729
|
|
GO:0023033
|
signaling pathway
|
0.491079 |
0.491079 |
2
|
956
|
|
GO:0042803
|
protein homodimerization activity
|
0.494624 |
0.494624 |
1
|
46
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.496927 |
0.496927 |
1
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.498346 |
0.498346 |
1
|
1959
|
|
GO:0035556
|
intracellular signal transduction
|
0.499208 |
0.499208 |
1
|
315
|
|
GO:0007399
|
nervous system development
|
0.500000 |
0.500000 |
1
|
848
|
|
GO:0043067
|
regulation of programmed cell death
|
0.509960 |
0.509960 |
1
|
128
|
|
GO:0048870
|
cell motility
|
0.512097 |
0.512097 |
1
|
254
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.513595 |
0.513595 |
1
|
850
|
|
GO:0044249
|
cellular biosynthetic process
|
0.526807 |
0.526807 |
1
|
2034
|
|
GO:0032501
|
multicellular organismal process
|
0.527040 |
0.527040 |
1
|
3315
|
|
GO:0010467
|
gene expression
|
0.531494 |
0.531494 |
1
|
1907
|
|
GO:0007265
|
Ras protein signal transduction
|
0.540541 |
0.540541 |
1
|
100
|
|
GO:0048592
|
eye morphogenesis
|
0.542069 |
0.542069 |
1
|
393
|
|
GO:0005737
|
cytoplasm
|
0.542427 |
0.542427 |
1
|
2378
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0035282
|
segmentation
|
0.581028 |
0.581028 |
1
|
294
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0007264
|
small GTPase mediated signal transduction
|
0.587302 |
0.587302 |
1
|
185
|
|
GO:0003702
|
RNA polymerase II transcription factor activity
|
0.618056 |
0.618056 |
1
|
267
|
|
GO:0006915
|
apoptosis
|
0.633333 |
0.633333 |
1
|
152
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.639129 |
0.639129 |
1
|
2878
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0042981
|
regulation of apoptosis
|
0.674699 |
0.674699 |
1
|
112
|
|
GO:0023034
|
intracellular signaling pathway
|
0.676453 |
0.676453 |
1
|
412
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.684327 |
0.684327 |
1
|
620
|
|
GO:0045449
|
regulation of transcription
|
0.687166 |
0.687166 |
1
|
771
|
|
GO:0043170
|
macromolecule metabolic process
|
0.690797 |
0.690797 |
1
|
3588
|
|
GO:0005623
|
cell
|
0.702620 |
0.702620 |
1
|
6195
|
|
GO:0044464
|
cell part
|
0.702620 |
0.702620 |
1
|
6195
|
|
GO:0044424
|
intracellular part
|
0.707667 |
0.707667 |
1
|
4384
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.724105 |
0.724105 |
1
|
1517
|
|
GO:0007292
|
female gamete generation
|
0.727380 |
0.727380 |
1
|
619
|
|
GO:0008152
|
metabolic process
|
0.739170 |
0.739170 |
1
|
5194
|
|
GO:0044237
|
cellular metabolic process
|
0.740940 |
0.740940 |
1
|
3814
|
|
GO:0005622
|
intracellular
|
0.764165 |
0.764165 |
1
|
4734
|
|
GO:0008361
|
regulation of cell size
|
0.769231 |
0.769231 |
1
|
100
|
|
GO:0043229
|
intracellular organelle
|
0.804100 |
0.804100 |
1
|
3530
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0001745
|
compound eye morphogenesis
|
0.811947 |
0.811947 |
1
|
367
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.814170 |
0.814170 |
1
|
701
|
|
GO:0044238
|
primary metabolic process
|
0.819985 |
0.819985 |
1
|
4259
|
|
GO:0007435
|
salivary gland morphogenesis
|
0.833333 |
0.833333 |
1
|
135
|
|
GO:0031267
|
small GTPase binding
|
0.833333 |
0.833333 |
1
|
25
|
|
GO:0001654
|
eye development
|
0.841912 |
0.841912 |
1
|
458
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.842767 |
0.842767 |
1
|
402
|
|
GO:0007431
|
salivary gland development
|
0.848168 |
0.848168 |
1
|
162
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0019953
|
sexual reproduction
|
0.859100 |
0.859100 |
1
|
878
|
|
GO:0007166
|
cell surface receptor linked signaling pathway
|
0.879634 |
0.879634 |
1
|
624
|
|
GO:0007276
|
gamete generation
|
0.883697 |
0.883697 |
1
|
851
|
|
GO:0008150
|
biological_process
|
0.925522 |
0.925522 |
2
|
10616
|
|
GO:0048749
|
compound eye development
|
0.932314 |
0.932314 |
1
|
427
|
|
GO:0035071
|
salivary gland cell autophagic cell death
|
0.938272 |
0.938272 |
1
|
76
|
|
GO:0012501
|
programmed cell death
|
0.941176 |
0.941176 |
1
|
240
|
|
GO:0003002
|
regionalization
|
0.942272 |
0.942272 |
1
|
506
|
|
GO:0002165
|
instar larval or pupal development
|
0.954000 |
0.954000 |
1
|
477
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0016477
|
cell migration
|
0.964567 |
0.964567 |
1
|
245
|
|
GO:0005575
|
cellular_component
|
0.970710 |
0.970710 |
1
|
8817
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0048477
|
oogenesis
|
0.987076 |
0.987076 |
1
|
611
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0017016
|
Ras GTPase binding
|
1.00000 |
1.00000 |
1
|
25
|
|
GO:0003700
|
sequence-specific DNA binding transcription factor activity
|
1.00000 |
1.00000 |
1
|
395
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|