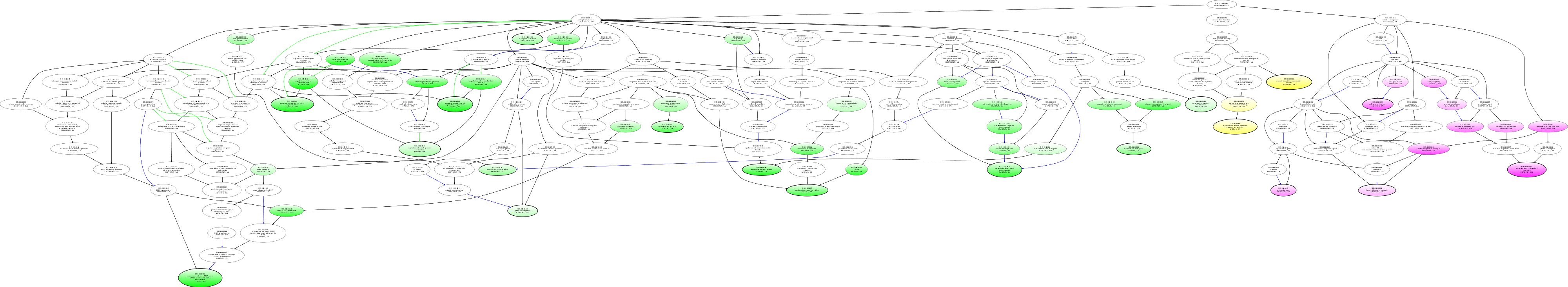
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0050792
|
regulation of viral reproduction
|
0.00177857 |
0.00177857 |
1
|
4
|
|
GO:0033168
|
conversion of ds siRNA to ss siRNA involved in RNA interference
|
0.00241546 |
0.00241546 |
1
|
1
|
|
GO:0016032
|
viral reproduction
|
0.00263529 |
0.00263529 |
1
|
7
|
|
GO:2000242
|
negative regulation of reproductive process
|
0.00394477 |
0.00394477 |
1
|
6
|
|
GO:0048525
|
negative regulation of viral reproduction
|
0.00564175 |
0.00564175 |
1
|
4
|
|
GO:0022415
|
viral reproductive process
|
0.00586510 |
0.00586510 |
1
|
6
|
|
GO:0001504
|
neurotransmitter uptake
|
0.00657895 |
0.00657895 |
1
|
1
|
|
GO:0005834
|
heterotrimeric G-protein complex
|
0.0127389 |
0.0127389 |
1
|
20
|
|
GO:0060004
|
reflex
|
0.0160000 |
0.0160000 |
1
|
6
|
|
GO:2000241
|
regulation of reproductive process
|
0.0166963 |
0.0166963 |
1
|
47
|
|
GO:0035050
|
embryonic heart tube development
|
0.0236111 |
0.0236111 |
1
|
17
|
|
GO:0031050
|
dsRNA fragmentation
|
0.0241546 |
0.0241546 |
1
|
10
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.0266285 |
0.0266285 |
3
|
2108
|
|
GO:0071705
|
nitrogen compound transport
|
0.0287241 |
0.0287241 |
1
|
43
|
|
GO:0007637
|
proboscis extension reflex
|
0.0340909 |
0.0340909 |
1
|
6
|
|
GO:0030529
|
ribonucleoprotein complex
|
0.0344737 |
0.0344737 |
2
|
510
|
|
GO:0005326
|
neurotransmitter transporter activity
|
0.0371901 |
0.0371901 |
1
|
27
|
|
GO:0046685
|
response to arsenic
|
0.0384615 |
0.0384615 |
1
|
1
|
|
GO:0044463
|
cell projection part
|
0.0388179 |
0.0388179 |
1
|
61
|
|
GO:0007584
|
response to nutrient
|
0.0429727 |
0.0429727 |
1
|
14
|
|
GO:0065007
|
biological regulation
|
0.0453190 |
0.0453190 |
3
|
2548
|
|
GO:0072359
|
circulatory system development
|
0.0454009 |
0.0454009 |
1
|
77
|
|
GO:0072358
|
cardiovascular system development
|
0.0454009 |
0.0454009 |
1
|
77
|
|
GO:0032991
|
macromolecular complex
|
0.0474134 |
0.0474134 |
3
|
2151
|
|
GO:0035295
|
tube development
|
0.0515704 |
0.0515704 |
1
|
133
|
|
GO:0008504
|
monoamine transmembrane transporter activity
|
0.0517241 |
0.0517241 |
1
|
3
|
|
GO:0044459
|
plasma membrane part
|
0.0559004 |
0.0559004 |
2
|
373
|
|
GO:0071702
|
organic substance transport
|
0.0607882 |
0.0607882 |
1
|
91
|
|
GO:0007507
|
heart development
|
0.0619968 |
0.0619968 |
1
|
77
|
|
GO:0071944
|
cell periphery
|
0.0696819 |
0.0696819 |
2
|
724
|
|
GO:0015844
|
monoamine transport
|
0.0697674 |
0.0697674 |
1
|
3
|
|
GO:0043331
|
response to dsRNA
|
0.0724638 |
0.0724638 |
1
|
10
|
|
GO:0044445
|
cytosolic part
|
0.0730005 |
0.0730005 |
1
|
136
|
|
GO:0019898
|
extrinsic to membrane
|
0.0730341 |
0.0730341 |
1
|
52
|
|
GO:0008283
|
cell proliferation
|
0.0732651 |
0.0732651 |
1
|
200
|
|
GO:0022610
|
biological adhesion
|
0.0753989 |
0.0753989 |
1
|
206
|
|
GO:0007405
|
neuroblast proliferation
|
0.0762852 |
0.0762852 |
1
|
46
|
|
GO:0010035
|
response to inorganic substance
|
0.0806486 |
0.0806486 |
1
|
26
|
|
GO:0023052
|
signaling
|
0.0828658 |
0.0828658 |
2
|
1364
|
|
GO:0005275
|
amine transmembrane transporter activity
|
0.0845481 |
0.0845481 |
1
|
58
|
|
GO:0045069
|
regulation of viral genome replication
|
0.0851064 |
0.0851064 |
1
|
4
|
|
GO:0005886
|
plasma membrane
|
0.0851857 |
0.0851857 |
2
|
641
|
|
GO:0042995
|
cell projection
|
0.0892053 |
0.0892053 |
1
|
143
|
|
GO:0015934
|
large ribosomal subunit
|
0.0932771 |
0.0932771 |
1
|
106
|
|
GO:0016458
|
gene silencing
|
0.0957538 |
0.0957538 |
1
|
163
|
|
GO:0009991
|
response to extracellular stimulus
|
0.0960000 |
0.0960000 |
1
|
36
|
|
GO:0051231
|
spindle elongation
|
0.0968523 |
0.0968523 |
1
|
80
|
|
GO:0008105
|
asymmetric protein localization
|
0.0971129 |
0.0971129 |
1
|
37
|
|
GO:0006836
|
neurotransmitter transport
|
0.0975284 |
0.0975284 |
1
|
146
|
|
GO:0050896
|
response to stimulus
|
0.104095 |
0.104095 |
2
|
1548
|
|
GO:0030029
|
actin filament-based process
|
0.108705 |
0.108705 |
1
|
186
|
|
GO:0007155
|
cell adhesion
|
0.111846 |
0.111846 |
1
|
192
|
|
GO:0055059
|
asymmetric neuroblast division
|
0.117647 |
0.117647 |
1
|
30
|
|
GO:0005198
|
structural molecule activity
|
0.119376 |
0.119376 |
1
|
459
|
|
GO:0051301
|
cell division
|
0.119842 |
0.119842 |
1
|
206
|
|
GO:0005840
|
ribosome
|
0.131663 |
0.131663 |
1
|
194
|
|
GO:0010608
|
posttranscriptional regulation of gene expression
|
0.133607 |
0.133607 |
1
|
130
|
|
GO:0009892
|
negative regulation of metabolic process
|
0.133617 |
0.133617 |
1
|
372
|
|
GO:0070887
|
cellular response to chemical stimulus
|
0.137974 |
0.137974 |
1
|
65
|
|
GO:0060089
|
molecular transducer activity
|
0.144056 |
0.144056 |
1
|
559
|
|
GO:0009987
|
cellular process
|
0.145754 |
0.145754 |
4
|
6560
|
|
GO:0051179
|
localization
|
0.148830 |
0.148830 |
2
|
1896
|
|
GO:0016441
|
posttranscriptional gene silencing
|
0.160000 |
0.160000 |
1
|
44
|
|
GO:0042221
|
response to chemical stimulus
|
0.166527 |
0.166527 |
2
|
632
|
|
GO:0040029
|
regulation of gene expression, epigenetic
|
0.181912 |
0.181912 |
1
|
177
|
|
GO:0019897
|
extrinsic to plasma membrane
|
0.183652 |
0.183652 |
1
|
37
|
|
GO:0010605
|
negative regulation of macromolecule metabolic process
|
0.187925 |
0.187925 |
1
|
356
|
|
GO:0015294
|
solute:cation symporter activity
|
0.190244 |
0.190244 |
1
|
78
|
|
GO:0016044
|
cellular membrane organization
|
0.193867 |
0.193867 |
1
|
344
|
|
GO:0019226
|
transmission of nerve impulse
|
0.194826 |
0.194826 |
1
|
241
|
|
GO:0051716
|
cellular response to stimulus
|
0.195075 |
0.195075 |
1
|
374
|
|
GO:0065008
|
regulation of biological quality
|
0.198629 |
0.198629 |
2
|
729
|
|
GO:0045165
|
cell fate commitment
|
0.199843 |
0.199843 |
1
|
255
|
|
GO:0005887
|
integral to plasma membrane
|
0.203593 |
0.203593 |
1
|
204
|
|
GO:0007010
|
cytoskeleton organization
|
0.205424 |
0.205424 |
2
|
536
|
|
GO:0070918
|
production of small RNA involved in gene silencing by RNA
|
0.208333 |
0.208333 |
1
|
10
|
|
GO:0005215
|
transporter activity
|
0.213704 |
0.213704 |
1
|
852
|
|
GO:0044425
|
membrane part
|
0.221352 |
0.221352 |
2
|
1398
|
|
GO:0044306
|
neuron projection terminus
|
0.223301 |
0.223301 |
1
|
23
|
|
GO:0005829
|
cytosol
|
0.228127 |
0.228127 |
1
|
425
|
|
GO:0016192
|
vesicle-mediated transport
|
0.228410 |
0.228410 |
1
|
430
|
|
GO:0005575
|
cellular_component
|
0.228855 |
0.228855 |
4
|
8817
|
|
GO:0061351
|
neural precursor cell proliferation
|
0.230000 |
0.230000 |
1
|
46
|
|
GO:0071359
|
cellular response to dsRNA
|
0.232558 |
0.232558 |
1
|
10
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.240586 |
0.240586 |
2
|
1562
|
|
GO:0048519
|
negative regulation of biological process
|
0.240665 |
0.240665 |
1
|
706
|
|
GO:0003924
|
GTPase activity
|
0.241796 |
0.241796 |
1
|
140
|
|
GO:0030422
|
production of siRNA involved in RNA interference
|
0.242424 |
0.242424 |
1
|
8
|
|
GO:0009790
|
embryo development
|
0.242987 |
0.242987 |
1
|
641
|
|
GO:0044446
|
intracellular organelle part
|
0.243036 |
0.243036 |
2
|
2162
|
|
GO:0045176
|
apical protein localization
|
0.243243 |
0.243243 |
1
|
9
|
|
GO:0005623
|
cell
|
0.243645 |
0.243645 |
4
|
6195
|
|
GO:0044464
|
cell part
|
0.243645 |
0.243645 |
4
|
6195
|
|
GO:0007017
|
microtubule-based process
|
0.246490 |
0.246490 |
1
|
448
|
|
GO:0007154
|
cell communication
|
0.252882 |
0.252882 |
1
|
461
|
|
GO:0044422
|
organelle part
|
0.256299 |
0.256299 |
2
|
2176
|
|
GO:0023046
|
signaling process
|
0.257083 |
0.257083 |
1
|
760
|
|
GO:0046873
|
metal ion transmembrane transporter activity
|
0.268293 |
0.268293 |
1
|
110
|
|
GO:0071310
|
cellular response to organic substance
|
0.270440 |
0.270440 |
1
|
43
|
|
GO:0005261
|
cation channel activity
|
0.272340 |
0.272340 |
1
|
128
|
|
GO:0010629
|
negative regulation of gene expression
|
0.276112 |
0.276112 |
1
|
288
|
|
GO:0022803
|
passive transmembrane transporter activity
|
0.276860 |
0.276860 |
1
|
201
|
|
GO:0010467
|
gene expression
|
0.282416 |
0.282416 |
2
|
1907
|
|
GO:0008360
|
regulation of cell shape
|
0.283845 |
0.283845 |
1
|
112
|
|
GO:0016071
|
mRNA metabolic process
|
0.284635 |
0.284635 |
1
|
339
|
|
GO:0007052
|
mitotic spindle organization
|
0.285513 |
0.285513 |
1
|
203
|
|
GO:0031047
|
gene silencing by RNA
|
0.294479 |
0.294479 |
1
|
48
|
|
GO:0005328
|
neurotransmitter:sodium symporter activity
|
0.298507 |
0.298507 |
1
|
20
|
|
GO:0033267
|
axon part
|
0.301205 |
0.301205 |
1
|
25
|
|
GO:0016817
|
hydrolase activity, acting on acid anhydrides
|
0.301217 |
0.301217 |
1
|
594
|
|
GO:0001505
|
regulation of neurotransmitter levels
|
0.304305 |
0.304305 |
1
|
129
|
|
GO:0005681
|
spliceosomal complex
|
0.307237 |
0.307237 |
1
|
180
|
|
GO:0022838
|
substrate-specific channel activity
|
0.307448 |
0.307448 |
1
|
194
|
|
GO:0007186
|
G-protein coupled receptor protein signaling pathway
|
0.318910 |
0.318910 |
1
|
199
|
|
GO:0031226
|
intrinsic to plasma membrane
|
0.327614 |
0.327614 |
1
|
208
|
|
GO:0000003
|
reproduction
|
0.332995 |
0.332995 |
1
|
1022
|
|
GO:0022414
|
reproductive process
|
0.332995 |
0.332995 |
1
|
1022
|
|
GO:0006837
|
serotonin transport
|
0.333333 |
0.333333 |
1
|
1
|
|
GO:0015222
|
serotonin transmembrane transporter activity
|
0.333333 |
0.333333 |
1
|
1
|
|
GO:0007635
|
chemosensory behavior
|
0.338282 |
0.338282 |
1
|
176
|
|
GO:0017145
|
stem cell division
|
0.339806 |
0.339806 |
1
|
70
|
|
GO:0007267
|
cell-cell signaling
|
0.342066 |
0.342066 |
1
|
263
|
|
GO:0022402
|
cell cycle process
|
0.343522 |
0.343522 |
1
|
655
|
|
GO:0015081
|
sodium ion transmembrane transporter activity
|
0.355769 |
0.355769 |
1
|
74
|
|
GO:0003008
|
system process
|
0.360531 |
0.360531 |
1
|
664
|
|
GO:0007051
|
spindle organization
|
0.368421 |
0.368421 |
1
|
238
|
|
GO:0005216
|
ion channel activity
|
0.369141 |
0.369141 |
1
|
189
|
|
GO:0032501
|
multicellular organismal process
|
0.369984 |
0.369984 |
2
|
3315
|
|
GO:0019222
|
regulation of metabolic process
|
0.376562 |
0.376562 |
1
|
1242
|
|
GO:0006412
|
translation
|
0.382593 |
0.382593 |
1
|
600
|
|
GO:0006396
|
RNA processing
|
0.382740 |
0.382740 |
1
|
414
|
|
GO:0007049
|
cell cycle
|
0.387308 |
0.387308 |
1
|
756
|
|
GO:0000022
|
mitotic spindle elongation
|
0.389163 |
0.389163 |
1
|
79
|
|
GO:0010033
|
response to organic substance
|
0.389301 |
0.389301 |
1
|
138
|
|
GO:0022625
|
cytosolic large ribosomal subunit
|
0.391892 |
0.391892 |
1
|
58
|
|
GO:0003735
|
structural constituent of ribosome
|
0.398693 |
0.398693 |
1
|
183
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.408435 |
0.408435 |
2
|
2878
|
|
GO:0016020
|
membrane
|
0.423766 |
0.423766 |
2
|
2122
|
|
GO:0061024
|
membrane organization
|
0.424507 |
0.424507 |
1
|
345
|
|
GO:0009605
|
response to external stimulus
|
0.425931 |
0.425931 |
1
|
375
|
|
GO:0051780
|
behavioral response to nutrient
|
0.428571 |
0.428571 |
1
|
6
|
|
GO:0022626
|
cytosolic ribosome
|
0.434783 |
0.434783 |
1
|
100
|
|
GO:0022008
|
neurogenesis
|
0.452398 |
0.452398 |
1
|
632
|
|
GO:0015291
|
secondary active transmembrane transporter activity
|
0.458333 |
0.458333 |
1
|
165
|
|
GO:0051234
|
establishment of localization
|
0.467930 |
0.467930 |
1
|
1549
|
|
GO:0044428
|
nuclear part
|
0.469235 |
0.469235 |
1
|
830
|
|
GO:0015837
|
amine transport
|
0.472527 |
0.472527 |
1
|
43
|
|
GO:0033036
|
macromolecule localization
|
0.476467 |
0.476467 |
1
|
524
|
|
GO:0043170
|
macromolecule metabolic process
|
0.477159 |
0.477159 |
2
|
3588
|
|
GO:0016787
|
hydrolase activity
|
0.480273 |
0.480273 |
1
|
1972
|
|
GO:0008150
|
biological_process
|
0.481028 |
0.481028 |
4
|
10616
|
|
GO:0015293
|
symporter activity
|
0.484848 |
0.484848 |
1
|
80
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.486014 |
0.486014 |
1
|
1056
|
|
GO:0022804
|
active transmembrane transporter activity
|
0.495868 |
0.495868 |
1
|
360
|
|
GO:0007399
|
nervous system development
|
0.500000 |
0.500000 |
1
|
848
|
|
GO:0022890
|
inorganic cation transmembrane transporter activity
|
0.502439 |
0.502439 |
1
|
206
|
|
GO:0043005
|
neuron projection
|
0.503497 |
0.503497 |
1
|
72
|
|
GO:0043226
|
organelle
|
0.526818 |
0.526818 |
2
|
3537
|
|
GO:0048869
|
cellular developmental process
|
0.540817 |
0.540817 |
1
|
1276
|
|
GO:0035195
|
gene silencing by miRNA
|
0.545455 |
0.545455 |
1
|
24
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0048103
|
somatic stem cell division
|
0.557143 |
0.557143 |
1
|
39
|
|
GO:0030036
|
actin cytoskeleton organization
|
0.564180 |
0.564180 |
1
|
183
|
|
GO:0000278
|
mitotic cell cycle
|
0.567460 |
0.567460 |
1
|
429
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.573530 |
0.573530 |
1
|
1227
|
|
GO:0055057
|
neuroblast division
|
0.574074 |
0.574074 |
1
|
31
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.574428 |
0.574428 |
1
|
1227
|
|
GO:0007610
|
behavior
|
0.591970 |
0.591970 |
1
|
559
|
|
GO:0007015
|
actin filament organization
|
0.606557 |
0.606557 |
1
|
111
|
|
GO:0006996
|
organelle organization
|
0.606991 |
0.606991 |
2
|
1182
|
|
GO:0050789
|
regulation of biological process
|
0.613640 |
0.613640 |
1
|
2246
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0009058
|
biosynthetic process
|
0.642905 |
0.642905 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.643595 |
0.643595 |
1
|
2093
|
|
GO:0043229
|
intracellular organelle
|
0.646541 |
0.646541 |
2
|
3530
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.646807 |
0.646807 |
1
|
1623
|
|
GO:0007166
|
cell surface receptor linked signaling pathway
|
0.652720 |
0.652720 |
1
|
624
|
|
GO:0006897
|
endocytosis
|
0.655814 |
0.655814 |
1
|
282
|
|
GO:0016070
|
RNA metabolic process
|
0.656488 |
0.656488 |
1
|
1191
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.658716 |
0.658716 |
1
|
1789
|
|
GO:0019079
|
viral genome replication
|
0.666667 |
0.666667 |
1
|
4
|
|
GO:0045071
|
negative regulation of viral genome replication
|
0.666667 |
0.666667 |
1
|
4
|
|
GO:0008380
|
RNA splicing
|
0.669082 |
0.669082 |
1
|
277
|
|
GO:0044444
|
cytoplasmic part
|
0.669378 |
0.669378 |
1
|
1863
|
|
GO:0008152
|
metabolic process
|
0.671256 |
0.671256 |
2
|
5194
|
|
GO:0044238
|
primary metabolic process
|
0.672346 |
0.672346 |
2
|
4259
|
|
GO:0044237
|
cellular metabolic process
|
0.673909 |
0.673909 |
2
|
3814
|
|
GO:0006911
|
phagocytosis, engulfment
|
0.680851 |
0.680851 |
1
|
192
|
|
GO:0032502
|
developmental process
|
0.681102 |
0.681102 |
1
|
2638
|
|
GO:0071013
|
catalytic step 2 spliceosome
|
0.683333 |
0.683333 |
1
|
123
|
|
GO:0044267
|
cellular protein metabolic process
|
0.686538 |
0.686538 |
1
|
1505
|
|
GO:0030424
|
axon
|
0.694444 |
0.694444 |
1
|
50
|
|
GO:0015370
|
solute:sodium symporter activity
|
0.701149 |
0.701149 |
1
|
61
|
|
GO:0016246
|
RNA interference
|
0.704545 |
0.704545 |
1
|
31
|
|
GO:0006909
|
phagocytosis
|
0.723404 |
0.723404 |
1
|
204
|
|
GO:0019538
|
protein metabolic process
|
0.724907 |
0.724907 |
1
|
2053
|
|
GO:0008104
|
protein localization
|
0.727099 |
0.727099 |
1
|
381
|
|
GO:0006397
|
mRNA processing
|
0.734411 |
0.734411 |
1
|
318
|
|
GO:0010468
|
regulation of gene expression
|
0.739954 |
0.739954 |
1
|
973
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.746994 |
0.746994 |
1
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.748407 |
0.748407 |
1
|
1959
|
|
GO:0003824
|
catalytic activity
|
0.751316 |
0.751316 |
1
|
4106
|
|
GO:0044249
|
cellular biosynthetic process
|
0.776153 |
0.776153 |
1
|
2034
|
|
GO:0000226
|
microtubule cytoskeleton organization
|
0.779196 |
0.779196 |
1
|
339
|
|
GO:0022891
|
substrate-specific transmembrane transporter activity
|
0.788146 |
0.788146 |
1
|
625
|
|
GO:0005737
|
cytoplasm
|
0.790684 |
0.790684 |
1
|
2378
|
|
GO:0008324
|
cation transmembrane transporter activity
|
0.808679 |
0.808679 |
1
|
410
|
|
GO:0015075
|
ion transmembrane transporter activity
|
0.811200 |
0.811200 |
1
|
507
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.813775 |
0.813775 |
2
|
1517
|
|
GO:0022892
|
substrate-specific transporter activity
|
0.815728 |
0.815728 |
1
|
695
|
|
GO:0010324
|
membrane invagination
|
0.819767 |
0.819767 |
1
|
282
|
|
GO:0071011
|
precatalytic spliceosome
|
0.822222 |
0.822222 |
1
|
148
|
|
GO:0000398
|
nuclear mRNA splicing, via spliceosome
|
0.836478 |
0.836478 |
1
|
266
|
|
GO:0015077
|
monovalent inorganic cation transmembrane transporter activity
|
0.844660 |
0.844660 |
1
|
174
|
|
GO:0022857
|
transmembrane transporter activity
|
0.852113 |
0.852113 |
1
|
726
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0007268
|
synaptic transmission
|
0.866667 |
0.866667 |
1
|
234
|
|
GO:0007275
|
multicellular organismal development
|
0.887792 |
0.887792 |
1
|
2296
|
|
GO:0022403
|
cell cycle phase
|
0.896183 |
0.896183 |
1
|
587
|
|
GO:0023033
|
signaling pathway
|
0.910681 |
0.910681 |
1
|
956
|
|
GO:0003674
|
molecular_function
|
0.912936 |
0.912936 |
3
|
11064
|
|
GO:0035194
|
posttranscriptional gene silencing by RNA
|
0.916667 |
0.916667 |
1
|
44
|
|
GO:0043679
|
axon terminus
|
0.920000 |
0.920000 |
1
|
23
|
|
GO:0044424
|
intracellular part
|
0.922046 |
0.922046 |
2
|
4384
|
|
GO:0016043
|
cellular component organization
|
0.922366 |
0.922366 |
3
|
2052
|
|
GO:0031224
|
intrinsic to membrane
|
0.924695 |
0.924695 |
1
|
1014
|
|
GO:0000279
|
M phase
|
0.945486 |
0.945486 |
1
|
555
|
|
GO:0050877
|
neurological system process
|
0.953313 |
0.953313 |
1
|
633
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0005622
|
intracellular
|
0.956866 |
0.956866 |
2
|
4734
|
|
GO:0043227
|
membrane-bounded organelle
|
0.963300 |
0.963300 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.963413 |
0.963413 |
1
|
2856
|
|
GO:0000375
|
RNA splicing, via transesterification reactions
|
0.963899 |
0.963899 |
1
|
267
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0043234
|
protein complex
|
0.979752 |
0.979752 |
1
|
1564
|
|
GO:0017111
|
nucleoside-triphosphatase activity
|
0.983022 |
0.983022 |
1
|
579
|
|
GO:0016021
|
integral to membrane
|
0.984221 |
0.984221 |
1
|
998
|
|
GO:0016462
|
pyrophosphatase activity
|
0.991582 |
0.991582 |
1
|
589
|
|
GO:0000377
|
RNA splicing, via transesterification reactions with bulged adenosine as nucleophile
|
0.996255 |
0.996255 |
1
|
266
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
4
|
12747
|
|
GO:0051610
|
serotonin uptake
|
1.00000 |
1.00000 |
1
|
1
|
|
GO:0016818
|
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
|
1.00000 |
1.00000 |
1
|
594
|
|
GO:0019058
|
viral infectious cycle
|
1.00000 |
1.00000 |
1
|
6
|
|
GO:0004871
|
signal transducer activity
|
1.00000 |
1.00000 |
1
|
559
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|
|
GO:0015267
|
channel activity
|
1.00000 |
1.00000 |
1
|
201
|
|
GO:0031667
|
response to nutrient levels
|
1.00000 |
1.00000 |
1
|
36
|