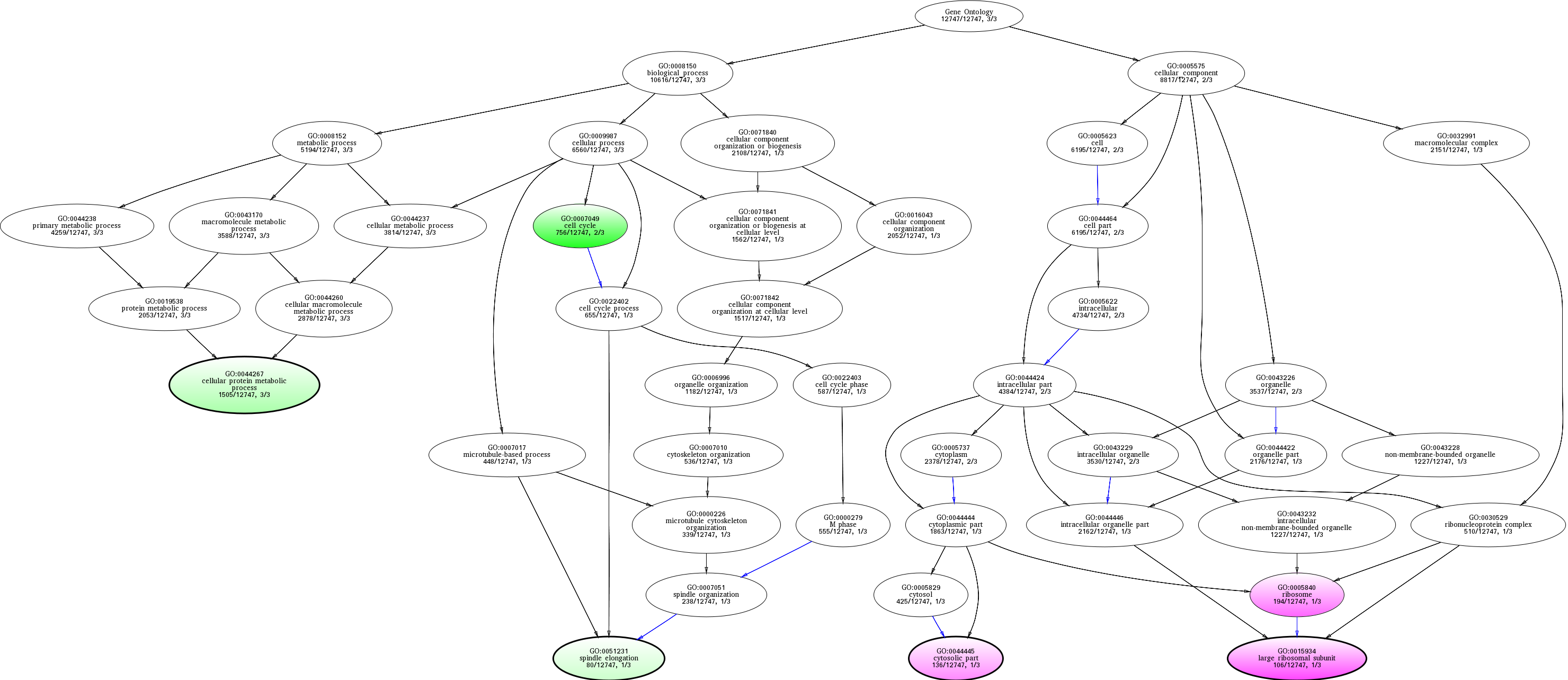
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0007049
|
cell cycle
|
0.0367464 |
0.0367464 |
2
|
756
|
|
GO:0015934
|
large ribosomal subunit
|
0.0477693 |
0.0477693 |
1
|
106
|
|
GO:0005840
|
ribosome
|
0.0681419 |
0.0681419 |
1
|
194
|
|
GO:0044445
|
cytosolic part
|
0.0730005 |
0.0730005 |
1
|
136
|
|
GO:0044267
|
cellular protein metabolic process
|
0.0851228 |
0.0851228 |
3
|
1505
|
|
GO:0051231
|
spindle elongation
|
0.0968523 |
0.0968523 |
1
|
80
|
|
GO:0043412
|
macromolecule modification
|
0.105638 |
0.105638 |
2
|
724
|
|
GO:0019538
|
protein metabolic process
|
0.107395 |
0.107395 |
3
|
2053
|
|
GO:0016874
|
ligase activity
|
0.109366 |
0.109366 |
1
|
231
|
|
GO:0008152
|
metabolic process
|
0.117083 |
0.117083 |
3
|
5194
|
|
GO:0044237
|
cellular metabolic process
|
0.118315 |
0.118315 |
3
|
3814
|
|
GO:0005198
|
structural molecule activity
|
0.119376 |
0.119376 |
1
|
459
|
|
GO:0005811
|
lipid particle
|
0.134192 |
0.134192 |
1
|
250
|
|
GO:0043226
|
organelle
|
0.160900 |
0.160900 |
2
|
3537
|
|
GO:0007017
|
microtubule-based process
|
0.191232 |
0.191232 |
1
|
448
|
|
GO:0070647
|
protein modification by small protein conjugation or removal
|
0.196964 |
0.196964 |
1
|
71
|
|
GO:0030529
|
ribonucleoprotein complex
|
0.210589 |
0.210589 |
1
|
510
|
|
GO:0006396
|
RNA processing
|
0.214286 |
0.214286 |
1
|
414
|
|
GO:0016311
|
dephosphorylation
|
0.221223 |
0.221223 |
1
|
123
|
|
GO:0005829
|
cytosol
|
0.228127 |
0.228127 |
1
|
425
|
|
GO:0009987
|
cellular process
|
0.235914 |
0.235914 |
3
|
6560
|
|
GO:0006470
|
protein dephosphorylation
|
0.246454 |
0.246454 |
1
|
94
|
|
GO:0016788
|
hydrolase activity, acting on ester bonds
|
0.251521 |
0.251521 |
1
|
496
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.260978 |
0.260978 |
3
|
2878
|
|
GO:0022402
|
cell cycle process
|
0.270667 |
0.270667 |
1
|
655
|
|
GO:0005737
|
cytoplasm
|
0.294170 |
0.294170 |
2
|
2378
|
|
GO:0003824
|
catalytic activity
|
0.310936 |
0.310936 |
2
|
4106
|
|
GO:0000278
|
mitotic cell cycle
|
0.321686 |
0.321686 |
2
|
429
|
|
GO:0043170
|
macromolecule metabolic process
|
0.329564 |
0.329564 |
3
|
3588
|
|
GO:0003723
|
RNA binding
|
0.351482 |
0.351482 |
1
|
652
|
|
GO:0007051
|
spindle organization
|
0.368421 |
0.368421 |
1
|
238
|
|
GO:0006793
|
phosphorus metabolic process
|
0.376764 |
0.376764 |
1
|
556
|
|
GO:0000022
|
mitotic spindle elongation
|
0.389163 |
0.389163 |
1
|
79
|
|
GO:0022625
|
cytosolic large ribosomal subunit
|
0.391892 |
0.391892 |
1
|
58
|
|
GO:0003735
|
structural constituent of ribosome
|
0.398693 |
0.398693 |
1
|
183
|
|
GO:0006464
|
protein modification process
|
0.414823 |
0.414823 |
2
|
684
|
|
GO:0032991
|
macromolecular complex
|
0.428425 |
0.428425 |
1
|
2151
|
|
GO:0044422
|
organelle part
|
0.432705 |
0.432705 |
1
|
2176
|
|
GO:0004722
|
protein serine/threonine phosphatase activity
|
0.434343 |
0.434343 |
1
|
43
|
|
GO:0022626
|
cytosolic ribosome
|
0.434783 |
0.434783 |
1
|
100
|
|
GO:0042578
|
phosphoric ester hydrolase activity
|
0.441532 |
0.441532 |
1
|
219
|
|
GO:0007010
|
cytoskeleton organization
|
0.453469 |
0.453469 |
1
|
536
|
|
GO:0043167
|
ion binding
|
0.460626 |
0.460626 |
1
|
1498
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.485282 |
0.485282 |
1
|
2108
|
|
GO:0007052
|
mitotic spindle organization
|
0.489796 |
0.489796 |
1
|
203
|
|
GO:0005623
|
cell
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044424
|
intracellular part
|
0.500760 |
0.500760 |
2
|
4384
|
|
GO:0005488
|
binding
|
0.514777 |
0.514777 |
2
|
5641
|
|
GO:0006412
|
translation
|
0.514942 |
0.514942 |
1
|
600
|
|
GO:0004721
|
phosphoprotein phosphatase activity
|
0.523810 |
0.523810 |
1
|
99
|
|
GO:0000226
|
microtubule cytoskeleton organization
|
0.529687 |
0.529687 |
1
|
339
|
|
GO:0003676
|
nucleic acid binding
|
0.549587 |
0.549587 |
1
|
1855
|
|
GO:0044238
|
primary metabolic process
|
0.551267 |
0.551267 |
3
|
4259
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.556542 |
0.556542 |
1
|
1562
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.573530 |
0.573530 |
1
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.574428 |
0.574428 |
1
|
1227
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0005622
|
intracellular
|
0.583919 |
0.583919 |
2
|
4734
|
|
GO:0016879
|
ligase activity, forming carbon-nitrogen bonds
|
0.610390 |
0.610390 |
1
|
141
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0043229
|
intracellular organelle
|
0.646541 |
0.646541 |
2
|
3530
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0044444
|
cytoplasmic part
|
0.669378 |
0.669378 |
1
|
1863
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.724105 |
0.724105 |
1
|
1517
|
|
GO:0016787
|
hydrolase activity
|
0.729944 |
0.729944 |
1
|
1972
|
|
GO:0044446
|
intracellular organelle part
|
0.743053 |
0.743053 |
1
|
2162
|
|
GO:0019787
|
small conjugating protein ligase activity
|
0.754386 |
0.754386 |
1
|
86
|
|
GO:0032446
|
protein modification by small protein conjugation
|
0.760563 |
0.760563 |
1
|
54
|
|
GO:0005575
|
cellular_component
|
0.773470 |
0.773470 |
2
|
8817
|
|
GO:0006996
|
organelle organization
|
0.779169 |
0.779169 |
1
|
1182
|
|
GO:0009058
|
biosynthetic process
|
0.786651 |
0.786651 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.787269 |
0.787269 |
1
|
2093
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.790151 |
0.790151 |
1
|
1623
|
|
GO:0016070
|
RNA metabolic process
|
0.798742 |
0.798742 |
1
|
1191
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.800673 |
0.800673 |
1
|
1789
|
|
GO:0016881
|
acid-amino acid ligase activity
|
0.808511 |
0.808511 |
1
|
114
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0016791
|
phosphatase activity
|
0.863014 |
0.863014 |
1
|
189
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.872797 |
0.872797 |
1
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.873851 |
0.873851 |
1
|
1959
|
|
GO:0044249
|
cellular biosynthetic process
|
0.894138 |
0.894138 |
1
|
2034
|
|
GO:0022403
|
cell cycle phase
|
0.896183 |
0.896183 |
1
|
587
|
|
GO:0010467
|
gene expression
|
0.897261 |
0.897261 |
1
|
1907
|
|
GO:0016567
|
protein ubiquitination
|
0.944444 |
0.944444 |
1
|
51
|
|
GO:0000279
|
M phase
|
0.945486 |
0.945486 |
1
|
555
|
|
GO:0043227
|
membrane-bounded organelle
|
0.963300 |
0.963300 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.963413 |
0.963413 |
1
|
2856
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0004842
|
ubiquitin-protein ligase activity
|
0.976744 |
0.976744 |
1
|
84
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0006796
|
phosphate metabolic process
|
1.00000 |
1.00000 |
1
|
556
|