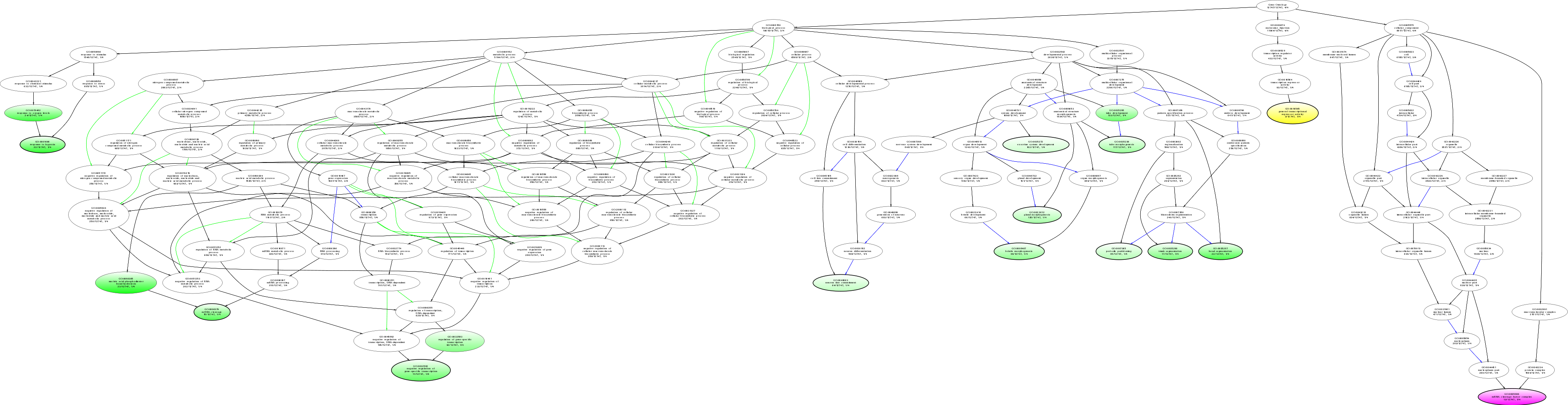
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0005849
|
mRNA cleavage factor complex
|
0.00805952 |
0.00805952 |
1
|
13
|
|
GO:0090305
|
nucleic acid phosphodiester bond hydrolysis
|
0.0297525 |
0.0297525 |
1
|
23
|
|
GO:0016565
|
general transcriptional repressor activity
|
0.0322581 |
0.0322581 |
1
|
3
|
|
GO:0001666
|
response to hypoxia
|
0.0363636 |
0.0363636 |
1
|
22
|
|
GO:0070482
|
response to oxygen levels
|
0.0379747 |
0.0379747 |
1
|
24
|
|
GO:0035287
|
head segmentation
|
0.0409683 |
0.0409683 |
1
|
22
|
|
GO:0006379
|
mRNA cleavage
|
0.0460123 |
0.0460123 |
1
|
15
|
|
GO:0032582
|
negative regulation of gene-specific transcription
|
0.0491071 |
0.0491071 |
1
|
11
|
|
GO:0035295
|
tube development
|
0.0515704 |
0.0515704 |
1
|
133
|
|
GO:0035290
|
trunk segmentation
|
0.0578231 |
0.0578231 |
1
|
17
|
|
GO:0032583
|
regulation of gene-specific transcription
|
0.0677419 |
0.0677419 |
1
|
42
|
|
GO:0008407
|
bristle morphogenesis
|
0.0706052 |
0.0706052 |
1
|
49
|
|
GO:0035239
|
tube morphogenesis
|
0.0759740 |
0.0759740 |
1
|
117
|
|
GO:0022612
|
gland morphogenesis
|
0.0873786 |
0.0873786 |
1
|
135
|
|
GO:0007365
|
periodic partitioning
|
0.0889328 |
0.0889328 |
1
|
45
|
|
GO:0048663
|
neuron fate commitment
|
0.0942563 |
0.0942563 |
1
|
64
|
|
GO:0035272
|
exocrine system development
|
0.0955189 |
0.0955189 |
1
|
162
|
|
GO:0016044
|
cellular membrane organization
|
0.102136 |
0.102136 |
1
|
344
|
|
GO:0003676
|
nucleic acid binding
|
0.108098 |
0.108098 |
2
|
1855
|
|
GO:0022416
|
bristle development
|
0.121324 |
0.121324 |
1
|
66
|
|
GO:0009890
|
negative regulation of biosynthetic process
|
0.129715 |
0.129715 |
1
|
282
|
|
GO:0009892
|
negative regulation of metabolic process
|
0.133617 |
0.133617 |
1
|
372
|
|
GO:0031327
|
negative regulation of cellular biosynthetic process
|
0.136166 |
0.136166 |
1
|
282
|
|
GO:0045465
|
R8 cell differentiation
|
0.141791 |
0.141791 |
1
|
19
|
|
GO:0000122
|
negative regulation of transcription from RNA polymerase II promoter
|
0.145455 |
0.145455 |
1
|
56
|
|
GO:0060541
|
respiratory system development
|
0.146816 |
0.146816 |
1
|
249
|
|
GO:0030528
|
transcription regulator activity
|
0.147289 |
0.147289 |
1
|
432
|
|
GO:0031324
|
negative regulation of cellular metabolic process
|
0.152355 |
0.152355 |
1
|
319
|
|
GO:0048732
|
gland development
|
0.153784 |
0.153784 |
1
|
191
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.162334 |
0.162334 |
2
|
2093
|
|
GO:0010558
|
negative regulation of macromolecule biosynthetic process
|
0.165094 |
0.165094 |
1
|
280
|
|
GO:0061024
|
membrane organization
|
0.168129 |
0.168129 |
1
|
345
|
|
GO:0016070
|
RNA metabolic process
|
0.171170 |
0.171170 |
2
|
1191
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.172797 |
0.172797 |
2
|
1789
|
|
GO:2000113
|
negative regulation of cellular macromolecule biosynthetic process
|
0.172946 |
0.172946 |
1
|
280
|
|
GO:0007366
|
periodic partitioning by pair rule gene
|
0.177778 |
0.177778 |
1
|
8
|
|
GO:0048523
|
negative regulation of cellular process
|
0.183975 |
0.183975 |
1
|
635
|
|
GO:0048519
|
negative regulation of biological process
|
0.186553 |
0.186553 |
1
|
706
|
|
GO:0010605
|
negative regulation of macromolecule metabolic process
|
0.187925 |
0.187925 |
1
|
356
|
|
GO:0032989
|
cellular component morphogenesis
|
0.193108 |
0.193108 |
1
|
566
|
|
GO:0045165
|
cell fate commitment
|
0.199843 |
0.199843 |
1
|
255
|
|
GO:0007389
|
pattern specification process
|
0.203563 |
0.203563 |
1
|
537
|
|
GO:0016564
|
transcription repressor activity
|
0.215278 |
0.215278 |
1
|
93
|
|
GO:0051172
|
negative regulation of nitrogen compound metabolic process
|
0.216720 |
0.216720 |
1
|
250
|
|
GO:0005575
|
cellular_component
|
0.228855 |
0.228855 |
4
|
8817
|
|
GO:0044237
|
cellular metabolic process
|
0.241038 |
0.241038 |
2
|
3814
|
|
GO:0009790
|
embryo development
|
0.242987 |
0.242987 |
1
|
641
|
|
GO:0016481
|
negative regulation of transcription
|
0.243724 |
0.243724 |
1
|
233
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.248286 |
0.248286 |
2
|
1959
|
|
GO:0045934
|
negative regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.252452 |
0.252452 |
1
|
250
|
|
GO:0007460
|
R8 cell fate commitment
|
0.254237 |
0.254237 |
1
|
15
|
|
GO:0031974
|
membrane-enclosed lumen
|
0.262807 |
0.262807 |
1
|
647
|
|
GO:0045892
|
negative regulation of transcription, DNA-dependent
|
0.266030 |
0.266030 |
1
|
195
|
|
GO:0010629
|
negative regulation of gene expression
|
0.276112 |
0.276112 |
1
|
288
|
|
GO:0010467
|
gene expression
|
0.282416 |
0.282416 |
2
|
1907
|
|
GO:0006357
|
regulation of transcription from RNA polymerase II promoter
|
0.282934 |
0.282934 |
1
|
189
|
|
GO:0051253
|
negative regulation of RNA metabolic process
|
0.287333 |
0.287333 |
1
|
202
|
|
GO:0043233
|
organelle lumen
|
0.291360 |
0.291360 |
1
|
634
|
|
GO:0070013
|
intracellular organelle lumen
|
0.293247 |
0.293247 |
1
|
634
|
|
GO:0009880
|
embryonic pattern specification
|
0.294591 |
0.294591 |
1
|
256
|
|
GO:0046530
|
photoreceptor cell differentiation
|
0.295620 |
0.295620 |
1
|
162
|
|
GO:0044451
|
nucleoplasm part
|
0.313253 |
0.313253 |
1
|
260
|
|
GO:0048869
|
cellular developmental process
|
0.322329 |
0.322329 |
1
|
1276
|
|
GO:0005654
|
nucleoplasm
|
0.340964 |
0.340964 |
1
|
283
|
|
GO:0046552
|
photoreceptor cell fate commitment
|
0.345238 |
0.345238 |
1
|
58
|
|
GO:0001751
|
compound eye photoreceptor cell differentiation
|
0.354497 |
0.354497 |
1
|
134
|
|
GO:0006366
|
transcription from RNA polymerase II promoter
|
0.360913 |
0.360913 |
1
|
253
|
|
GO:0001754
|
eye photoreceptor cell differentiation
|
0.361386 |
0.361386 |
1
|
146
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.366791 |
0.366791 |
1
|
888
|
|
GO:0042706
|
eye photoreceptor cell fate commitment
|
0.371622 |
0.371622 |
1
|
55
|
|
GO:0009887
|
organ morphogenesis
|
0.374384 |
0.374384 |
1
|
684
|
|
GO:0019222
|
regulation of metabolic process
|
0.376562 |
0.376562 |
1
|
1242
|
|
GO:0050896
|
response to stimulus
|
0.376795 |
0.376795 |
1
|
1548
|
|
GO:0006396
|
RNA processing
|
0.382740 |
0.382740 |
1
|
414
|
|
GO:0006950
|
response to stress
|
0.390827 |
0.390827 |
1
|
605
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.396252 |
0.396252 |
1
|
888
|
|
GO:0016566
|
specific transcriptional repressor activity
|
0.397849 |
0.397849 |
1
|
37
|
|
GO:0042221
|
response to chemical stimulus
|
0.408269 |
0.408269 |
1
|
632
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.408435 |
0.408435 |
2
|
2878
|
|
GO:0005634
|
nucleus
|
0.408695 |
0.408695 |
2
|
1826
|
|
GO:0001752
|
compound eye photoreceptor fate commitment
|
0.410448 |
0.410448 |
1
|
55
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.418200 |
0.418200 |
1
|
1039
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.429576 |
0.429576 |
1
|
1110
|
|
GO:0007423
|
sensory organ development
|
0.438003 |
0.438003 |
1
|
544
|
|
GO:0030182
|
neuron differentiation
|
0.442292 |
0.442292 |
1
|
548
|
|
GO:0005622
|
intracellular
|
0.446165 |
0.446165 |
3
|
4734
|
|
GO:0022008
|
neurogenesis
|
0.452398 |
0.452398 |
1
|
632
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.456989 |
0.456989 |
1
|
850
|
|
GO:0043167
|
ion binding
|
0.460626 |
0.460626 |
1
|
1498
|
|
GO:0044428
|
nuclear part
|
0.469235 |
0.469235 |
1
|
830
|
|
GO:0031981
|
nuclear lumen
|
0.475277 |
0.475277 |
1
|
471
|
|
GO:0043170
|
macromolecule metabolic process
|
0.477159 |
0.477159 |
2
|
3588
|
|
GO:0016787
|
hydrolase activity
|
0.480273 |
0.480273 |
1
|
1972
|
|
GO:0008152
|
metabolic process
|
0.483893 |
0.483893 |
2
|
5194
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.485282 |
0.485282 |
1
|
2108
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.486014 |
0.486014 |
1
|
1056
|
|
GO:0016071
|
mRNA metabolic process
|
0.488424 |
0.488424 |
1
|
339
|
|
GO:0032774
|
RNA biosynthetic process
|
0.496008 |
0.496008 |
1
|
703
|
|
GO:0007399
|
nervous system development
|
0.500000 |
0.500000 |
1
|
848
|
|
GO:0050789
|
regulation of biological process
|
0.509927 |
0.509927 |
1
|
2246
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.513595 |
0.513595 |
1
|
850
|
|
GO:0005515
|
protein binding
|
0.518366 |
0.518366 |
1
|
1726
|
|
GO:0050794
|
regulation of cellular process
|
0.518911 |
0.518911 |
1
|
2034
|
|
GO:0043226
|
organelle
|
0.526818 |
0.526818 |
2
|
3537
|
|
GO:0048592
|
eye morphogenesis
|
0.542069 |
0.542069 |
1
|
393
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0065007
|
biological regulation
|
0.561089 |
0.561089 |
1
|
2548
|
|
GO:0003674
|
molecular_function
|
0.567526 |
0.567526 |
4
|
11064
|
|
GO:0032502
|
developmental process
|
0.575616 |
0.575616 |
1
|
2638
|
|
GO:0035282
|
segmentation
|
0.581028 |
0.581028 |
1
|
294
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.617799 |
0.617799 |
1
|
905
|
|
GO:0006350
|
transcription
|
0.625331 |
0.625331 |
1
|
859
|
|
GO:0009058
|
biosynthetic process
|
0.642905 |
0.642905 |
1
|
2090
|
|
GO:0043229
|
intracellular organelle
|
0.646541 |
0.646541 |
2
|
3530
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.646807 |
0.646807 |
1
|
1623
|
|
GO:0043227
|
membrane-bounded organelle
|
0.653325 |
0.653325 |
2
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.653801 |
0.653801 |
2
|
2856
|
|
GO:0005623
|
cell
|
0.656329 |
0.656329 |
3
|
6195
|
|
GO:0044464
|
cell part
|
0.656329 |
0.656329 |
3
|
6195
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0044238
|
primary metabolic process
|
0.672346 |
0.672346 |
2
|
4259
|
|
GO:0032991
|
macromolecular complex
|
0.673350 |
0.673350 |
1
|
2151
|
|
GO:0009987
|
cellular process
|
0.673638 |
0.673638 |
2
|
6560
|
|
GO:0032501
|
multicellular organismal process
|
0.674757 |
0.674757 |
1
|
3315
|
|
GO:0044422
|
organelle part
|
0.678224 |
0.678224 |
1
|
2176
|
|
GO:0035289
|
posterior head segmentation
|
0.681818 |
0.681818 |
1
|
15
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.684327 |
0.684327 |
1
|
620
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.686903 |
0.686903 |
1
|
686
|
|
GO:0045449
|
regulation of transcription
|
0.687166 |
0.687166 |
1
|
771
|
|
GO:0003677
|
DNA binding
|
0.691225 |
0.691225 |
1
|
824
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.699519 |
0.699519 |
1
|
903
|
|
GO:0005488
|
binding
|
0.702165 |
0.702165 |
2
|
5641
|
|
GO:0043234
|
protein complex
|
0.727104 |
0.727104 |
1
|
1564
|
|
GO:0006397
|
mRNA processing
|
0.734411 |
0.734411 |
1
|
318
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.736132 |
0.736132 |
2
|
1535
|
|
GO:0010468
|
regulation of gene expression
|
0.739954 |
0.739954 |
1
|
973
|
|
GO:0044446
|
intracellular organelle part
|
0.743053 |
0.743053 |
1
|
2162
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.746994 |
0.746994 |
1
|
1617
|
|
GO:0044249
|
cellular biosynthetic process
|
0.776153 |
0.776153 |
1
|
2034
|
|
GO:0007350
|
blastoderm segmentation
|
0.779221 |
0.779221 |
1
|
240
|
|
GO:0008270
|
zinc ion binding
|
0.781440 |
0.781440 |
1
|
901
|
|
GO:0044424
|
intracellular part
|
0.793631 |
0.793631 |
2
|
4384
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0001745
|
compound eye morphogenesis
|
0.811947 |
0.811947 |
1
|
367
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.814170 |
0.814170 |
1
|
701
|
|
GO:0000902
|
cell morphogenesis
|
0.825088 |
0.825088 |
1
|
467
|
|
GO:0007435
|
salivary gland morphogenesis
|
0.833333 |
0.833333 |
1
|
135
|
|
GO:0001654
|
eye development
|
0.841912 |
0.841912 |
1
|
458
|
|
GO:0003824
|
catalytic activity
|
0.843631 |
0.843631 |
1
|
4106
|
|
GO:0007431
|
salivary gland development
|
0.848168 |
0.848168 |
1
|
162
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0008150
|
biological_process
|
0.867374 |
0.867374 |
3
|
10616
|
|
GO:0048749
|
compound eye development
|
0.932314 |
0.932314 |
1
|
427
|
|
GO:0003002
|
regionalization
|
0.942272 |
0.942272 |
1
|
506
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0007424
|
open tracheal system development
|
0.995984 |
0.995984 |
1
|
248
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
4
|
12747
|