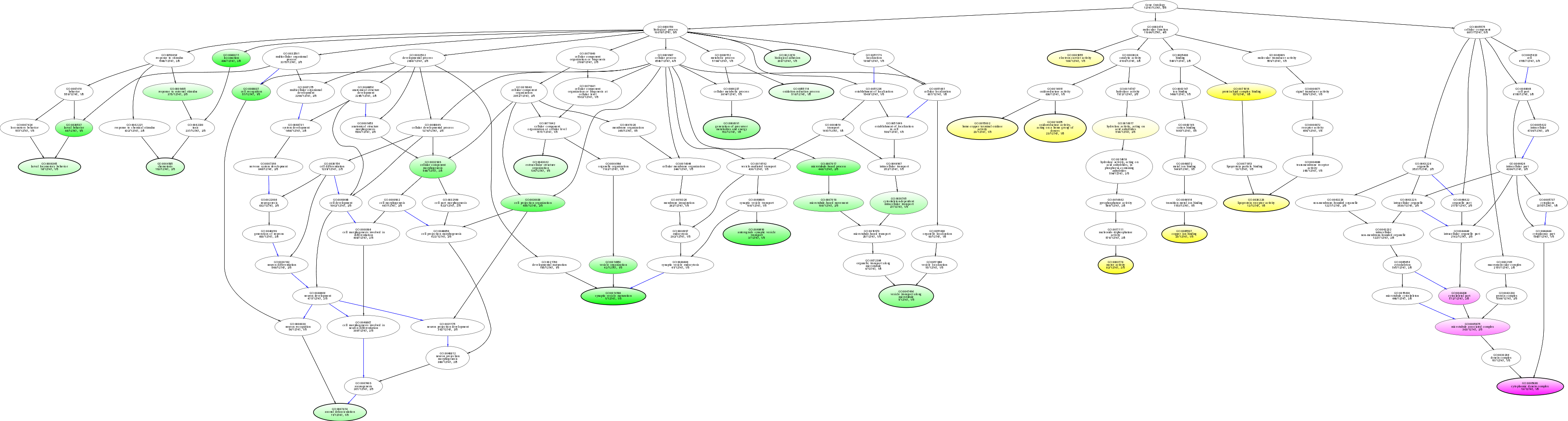
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0016188
|
synaptic vesicle maturation
|
0.00444444 |
0.00444444 |
1
|
1
|
|
GO:0071814
|
protein-lipid complex binding
|
0.00460421 |
0.00460421 |
1
|
13
|
|
GO:0005868
|
cytoplasmic dynein complex
|
0.00688559 |
0.00688559 |
1
|
13
|
|
GO:0040011
|
locomotion
|
0.0189241 |
0.0189241 |
2
|
484
|
|
GO:0003774
|
motor activity
|
0.0198469 |
0.0198469 |
2
|
82
|
|
GO:0005507
|
copper ion binding
|
0.0199480 |
0.0199480 |
1
|
23
|
|
GO:0007017
|
microtubule-based process
|
0.0254574 |
0.0254574 |
2
|
448
|
|
GO:0030537
|
larval behavior
|
0.0255469 |
0.0255469 |
1
|
44
|
|
GO:0048490
|
anterograde synaptic vesicle transport
|
0.0283019 |
0.0283019 |
1
|
3
|
|
GO:0008037
|
cell recognition
|
0.0293643 |
0.0293643 |
1
|
57
|
|
GO:0030030
|
cell projection organization
|
0.0295481 |
0.0295481 |
2
|
485
|
|
GO:0030228
|
lipoprotein receptor activity
|
0.0303030 |
0.0303030 |
1
|
12
|
|
GO:0016050
|
vesicle organization
|
0.0355330 |
0.0355330 |
1
|
42
|
|
GO:0016675
|
oxidoreductase activity, acting on a heme group of donors
|
0.0360502 |
0.0360502 |
1
|
23
|
|
GO:0015002
|
heme-copper terminal oxidase activity
|
0.0360502 |
0.0360502 |
1
|
23
|
|
GO:0032989
|
cellular component morphogenesis
|
0.0372376 |
0.0372376 |
2
|
566
|
|
GO:0006091
|
generation of precursor metabolites and energy
|
0.0398532 |
0.0398532 |
1
|
152
|
|
GO:0047496
|
vesicle transport along microtubule
|
0.0500000 |
0.0500000 |
1
|
1
|
|
GO:0005875
|
microtubule associated complex
|
0.0504126 |
0.0504126 |
2
|
363
|
|
GO:0044430
|
cytoskeletal part
|
0.0535924 |
0.0535924 |
2
|
512
|
|
GO:0007018
|
microtubule-based movement
|
0.0577061 |
0.0577061 |
2
|
108
|
|
GO:0009055
|
electron carrier activity
|
0.0579938 |
0.0579938 |
1
|
164
|
|
GO:0009605
|
response to external stimulus
|
0.0585655 |
0.0585655 |
2
|
375
|
|
GO:0030705
|
cytoskeleton-dependent intracellular transport
|
0.0767045 |
0.0767045 |
1
|
27
|
|
GO:0007414
|
axonal defasciculation
|
0.0796775 |
0.0796775 |
1
|
11
|
|
GO:0006935
|
chemotaxis
|
0.0824254 |
0.0824254 |
2
|
193
|
|
GO:0008345
|
larval locomotory behavior
|
0.0874317 |
0.0874317 |
1
|
16
|
|
GO:0043062
|
extracellular structure organization
|
0.0883322 |
0.0883322 |
1
|
134
|
|
GO:0016817
|
hydrolase activity, acting on acid anhydrides
|
0.0906249 |
0.0906249 |
2
|
594
|
|
GO:0022610
|
biological adhesion
|
0.0933472 |
0.0933472 |
1
|
206
|
|
GO:0055114
|
oxidation-reduction process
|
0.0993454 |
0.0993454 |
1
|
516
|
|
GO:0007155
|
cell adhesion
|
0.111846 |
0.111846 |
1
|
192
|
|
GO:0048488
|
synaptic vesicle endocytosis
|
0.120235 |
0.120235 |
1
|
41
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.120278 |
0.120278 |
2
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.120756 |
0.120756 |
2
|
1227
|
|
GO:0021700
|
developmental maturation
|
0.121205 |
0.121205 |
1
|
165
|
|
GO:0000904
|
cell morphogenesis involved in differentiation
|
0.141709 |
0.141709 |
2
|
408
|
|
GO:0022900
|
electron transport chain
|
0.143911 |
0.143911 |
1
|
78
|
|
GO:0005509
|
calcium ion binding
|
0.144237 |
0.144237 |
1
|
209
|
|
GO:0048869
|
cellular developmental process
|
0.146194 |
0.146194 |
2
|
1276
|
|
GO:0003824
|
catalytic activity
|
0.147500 |
0.147500 |
3
|
4106
|
|
GO:0007517
|
muscle organ development
|
0.149102 |
0.149102 |
1
|
191
|
|
GO:0042330
|
taxis
|
0.151554 |
0.151554 |
2
|
237
|
|
GO:0051640
|
organelle localization
|
0.153213 |
0.153213 |
1
|
93
|
|
GO:0050896
|
response to stimulus
|
0.157100 |
0.157100 |
2
|
1548
|
|
GO:0005575
|
cellular_component
|
0.158275 |
0.158275 |
5
|
8817
|
|
GO:0005871
|
kinesin complex
|
0.158669 |
0.158669 |
1
|
30
|
|
GO:0051648
|
vesicle localization
|
0.161290 |
0.161290 |
1
|
15
|
|
GO:0060537
|
muscle tissue development
|
0.163375 |
0.163375 |
1
|
91
|
|
GO:0042221
|
response to chemical stimulus
|
0.166527 |
0.166527 |
2
|
632
|
|
GO:0055001
|
muscle cell development
|
0.179057 |
0.179057 |
1
|
100
|
|
GO:0060089
|
molecular transducer activity
|
0.187314 |
0.187314 |
1
|
559
|
|
GO:0048666
|
neuron development
|
0.187871 |
0.187871 |
2
|
471
|
|
GO:0016044
|
cellular membrane organization
|
0.193867 |
0.193867 |
1
|
344
|
|
GO:0042692
|
muscle cell differentiation
|
0.194824 |
0.194824 |
1
|
132
|
|
GO:0030182
|
neuron differentiation
|
0.195423 |
0.195423 |
2
|
548
|
|
GO:0022008
|
neurogenesis
|
0.204486 |
0.204486 |
2
|
632
|
|
GO:0005856
|
cytoskeleton
|
0.208841 |
0.208841 |
2
|
561
|
|
GO:0048468
|
cell development
|
0.209123 |
0.209123 |
2
|
1042
|
|
GO:0061061
|
muscle structure development
|
0.210182 |
0.210182 |
1
|
252
|
|
GO:0030286
|
dynein complex
|
0.213415 |
0.213415 |
1
|
41
|
|
GO:0008038
|
neuron recognition
|
0.223434 |
0.223434 |
1
|
56
|
|
GO:0016198
|
axon choice point recognition
|
0.226131 |
0.226131 |
1
|
24
|
|
GO:0046907
|
intracellular transport
|
0.227832 |
0.227832 |
1
|
352
|
|
GO:0016192
|
vesicle-mediated transport
|
0.228410 |
0.228410 |
1
|
430
|
|
GO:0004129
|
cytochrome-c oxidase activity
|
0.230000 |
0.230000 |
1
|
23
|
|
GO:0005576
|
extracellular region
|
0.239639 |
0.239639 |
1
|
470
|
|
GO:0072384
|
organelle transport along microtubule
|
0.240000 |
0.240000 |
1
|
6
|
|
GO:0044446
|
intracellular organelle part
|
0.243036 |
0.243036 |
2
|
2162
|
|
GO:0048489
|
synaptic vesicle transport
|
0.246512 |
0.246512 |
1
|
106
|
|
GO:0007399
|
nervous system development
|
0.249853 |
0.249853 |
2
|
848
|
|
GO:0001882
|
nucleoside binding
|
0.258670 |
0.258670 |
1
|
784
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.259771 |
0.259771 |
2
|
2108
|
|
GO:0005215
|
transporter activity
|
0.274270 |
0.274270 |
1
|
852
|
|
GO:0007626
|
locomotory behavior
|
0.280859 |
0.280859 |
1
|
157
|
|
GO:0051649
|
establishment of localization in cell
|
0.303951 |
0.303951 |
1
|
500
|
|
GO:0061024
|
membrane organization
|
0.308058 |
0.308058 |
1
|
345
|
|
GO:0051641
|
cellular localization
|
0.309572 |
0.309572 |
1
|
607
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.338051 |
0.338051 |
2
|
1534
|
|
GO:0032991
|
macromolecular complex
|
0.354447 |
0.354447 |
2
|
2151
|
|
GO:0044422
|
organelle part
|
0.360428 |
0.360428 |
2
|
2176
|
|
GO:0032502
|
developmental process
|
0.364008 |
0.364008 |
2
|
2638
|
|
GO:0009987
|
cellular process
|
0.368587 |
0.368587 |
4
|
6560
|
|
GO:0000166
|
nucleotide binding
|
0.374077 |
0.374077 |
1
|
1178
|
|
GO:0016491
|
oxidoreductase activity
|
0.397549 |
0.397549 |
1
|
638
|
|
GO:0008150
|
biological_process
|
0.400586 |
0.400586 |
5
|
10616
|
|
GO:0010970
|
microtubule-based transport
|
0.404504 |
0.404504 |
1
|
25
|
|
GO:0009888
|
tissue development
|
0.431439 |
0.431439 |
1
|
557
|
|
GO:0048731
|
system development
|
0.432376 |
0.432376 |
2
|
1696
|
|
GO:0007275
|
multicellular organismal development
|
0.442066 |
0.442066 |
2
|
2296
|
|
GO:0043167
|
ion binding
|
0.460626 |
0.460626 |
1
|
1498
|
|
GO:0007409
|
axonogenesis
|
0.461014 |
0.461014 |
2
|
267
|
|
GO:0007411
|
axon guidance
|
0.466817 |
0.466817 |
2
|
186
|
|
GO:0016787
|
hydrolase activity
|
0.470417 |
0.470417 |
2
|
1972
|
|
GO:0031175
|
neuron projection development
|
0.484416 |
0.484416 |
2
|
392
|
|
GO:0032501
|
multicellular organismal process
|
0.496887 |
0.496887 |
2
|
3315
|
|
GO:0022890
|
inorganic cation transmembrane transporter activity
|
0.502439 |
0.502439 |
1
|
206
|
|
GO:0043234
|
protein complex
|
0.528587 |
0.528587 |
2
|
1564
|
|
GO:0044425
|
membrane part
|
0.535780 |
0.535780 |
1
|
1398
|
|
GO:0060538
|
skeletal muscle organ development
|
0.539267 |
0.539267 |
1
|
103
|
|
GO:0051234
|
establishment of localization
|
0.545594 |
0.545594 |
1
|
1549
|
|
GO:0032990
|
cell part morphogenesis
|
0.555558 |
0.555558 |
2
|
422
|
|
GO:0015078
|
hydrogen ion transmembrane transporter activity
|
0.574713 |
0.574713 |
1
|
100
|
|
GO:0007610
|
behavior
|
0.591970 |
0.591970 |
1
|
559
|
|
GO:0051179
|
localization
|
0.626157 |
0.626157 |
1
|
1896
|
|
GO:0048667
|
cell morphogenesis involved in neuron differentiation
|
0.632489 |
0.632489 |
2
|
389
|
|
GO:0007415
|
defasciculation of motor neuron axon
|
0.636364 |
0.636364 |
1
|
7
|
|
GO:0015630
|
microtubule cytoskeleton
|
0.637433 |
0.637433 |
2
|
448
|
|
GO:0043229
|
intracellular organelle
|
0.646541 |
0.646541 |
2
|
3530
|
|
GO:0048858
|
cell projection morphogenesis
|
0.650762 |
0.650762 |
2
|
422
|
|
GO:0006897
|
endocytosis
|
0.655814 |
0.655814 |
1
|
282
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.661861 |
0.661861 |
1
|
1562
|
|
GO:0043226
|
organelle
|
0.665095 |
0.665095 |
2
|
3537
|
|
GO:0044444
|
cytoplasmic part
|
0.669378 |
0.669378 |
1
|
1863
|
|
GO:0007519
|
skeletal muscle tissue development
|
0.669565 |
0.669565 |
1
|
77
|
|
GO:0000902
|
cell morphogenesis
|
0.680515 |
0.680515 |
2
|
467
|
|
GO:0007528
|
neuromuscular junction development
|
0.684783 |
0.684783 |
1
|
63
|
|
GO:0050808
|
synapse organization
|
0.686567 |
0.686567 |
1
|
92
|
|
GO:0005488
|
binding
|
0.702165 |
0.702165 |
2
|
5641
|
|
GO:0016020
|
membrane
|
0.715875 |
0.715875 |
1
|
2122
|
|
GO:0048747
|
muscle fiber development
|
0.717172 |
0.717172 |
1
|
71
|
|
GO:0031224
|
intrinsic to membrane
|
0.725322 |
0.725322 |
1
|
1014
|
|
GO:0048856
|
anatomical structure development
|
0.737157 |
0.737157 |
2
|
2265
|
|
GO:0032553
|
ribonucleotide binding
|
0.746180 |
0.746180 |
1
|
879
|
|
GO:0048741
|
skeletal muscle fiber development
|
0.750000 |
0.750000 |
1
|
63
|
|
GO:0032559
|
adenyl ribonucleotide binding
|
0.757705 |
0.757705 |
1
|
713
|
|
GO:0030169
|
low-density lipoprotein binding
|
0.769231 |
0.769231 |
1
|
10
|
|
GO:0006996
|
organelle organization
|
0.779169 |
0.779169 |
1
|
1182
|
|
GO:0022891
|
substrate-specific transmembrane transporter activity
|
0.788146 |
0.788146 |
1
|
625
|
|
GO:0005737
|
cytoplasm
|
0.790684 |
0.790684 |
1
|
2378
|
|
GO:0044424
|
intracellular part
|
0.793631 |
0.793631 |
2
|
4384
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0048513
|
organ development
|
0.796117 |
0.796117 |
1
|
1242
|
|
GO:0055002
|
striated muscle cell development
|
0.798387 |
0.798387 |
1
|
99
|
|
GO:0017076
|
purine nucleotide binding
|
0.803056 |
0.803056 |
1
|
946
|
|
GO:0008324
|
cation transmembrane transporter activity
|
0.808679 |
0.808679 |
1
|
410
|
|
GO:0015075
|
ion transmembrane transporter activity
|
0.811200 |
0.811200 |
1
|
507
|
|
GO:0022892
|
substrate-specific transporter activity
|
0.815728 |
0.815728 |
1
|
695
|
|
GO:0030554
|
adenyl nucleotide binding
|
0.819239 |
0.819239 |
1
|
775
|
|
GO:0010324
|
membrane invagination
|
0.819767 |
0.819767 |
1
|
282
|
|
GO:0004872
|
receptor activity
|
0.821109 |
0.821109 |
1
|
459
|
|
GO:0048812
|
neuron projection morphogenesis
|
0.829362 |
0.829362 |
2
|
388
|
|
GO:0005041
|
low-density lipoprotein receptor activity
|
0.833333 |
0.833333 |
1
|
10
|
|
GO:0005623
|
cell
|
0.840429 |
0.840429 |
3
|
6195
|
|
GO:0044464
|
cell part
|
0.840429 |
0.840429 |
3
|
6195
|
|
GO:0015077
|
monovalent inorganic cation transmembrane transporter activity
|
0.844660 |
0.844660 |
1
|
174
|
|
GO:0016887
|
ATPase activity
|
0.848056 |
0.848056 |
1
|
353
|
|
GO:0022857
|
transmembrane transporter activity
|
0.852113 |
0.852113 |
1
|
726
|
|
GO:0005622
|
intracellular
|
0.859425 |
0.859425 |
2
|
4734
|
|
GO:0004888
|
transmembrane receptor activity
|
0.860566 |
0.860566 |
1
|
395
|
|
GO:0003674
|
molecular_function
|
0.867344 |
0.867344 |
4
|
11064
|
|
GO:0003777
|
microtubule motor activity
|
0.869015 |
0.869015 |
1
|
52
|
|
GO:0042623
|
ATPase activity, coupled
|
0.900850 |
0.900850 |
1
|
318
|
|
GO:0048699
|
generation of neurons
|
0.910264 |
0.910264 |
2
|
603
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.923977 |
0.923977 |
1
|
1517
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.929175 |
0.929175 |
1
|
879
|
|
GO:0044237
|
cellular metabolic process
|
0.932921 |
0.932921 |
1
|
3814
|
|
GO:0051146
|
striated muscle cell differentiation
|
0.939394 |
0.939394 |
1
|
124
|
|
GO:0030154
|
cell differentiation
|
0.942825 |
0.942825 |
2
|
1239
|
|
GO:0016043
|
cellular component organization
|
0.947563 |
0.947563 |
2
|
2052
|
|
GO:0008152
|
metabolic process
|
0.965278 |
0.965278 |
1
|
5194
|
|
GO:0017111
|
nucleoside-triphosphatase activity
|
0.966304 |
0.966304 |
2
|
579
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0016462
|
pyrophosphatase activity
|
0.983222 |
0.983222 |
2
|
589
|
|
GO:0016021
|
integral to membrane
|
0.984221 |
0.984221 |
1
|
998
|
|
GO:0014706
|
striated muscle tissue development
|
0.989011 |
0.989011 |
1
|
90
|
|
GO:0001883
|
purine nucleoside binding
|
0.992347 |
0.992347 |
1
|
778
|
|
GO:0005524
|
ATP binding
|
0.998597 |
0.998597 |
1
|
712
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
5
|
12747
|
|
GO:0016818
|
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
|
1.00000 |
1.00000 |
2
|
594
|
|
GO:0016676
|
oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor
|
1.00000 |
1.00000 |
1
|
23
|
|
GO:0071813
|
lipoprotein particle binding
|
1.00000 |
1.00000 |
1
|
13
|
|
GO:0004871
|
signal transducer activity
|
1.00000 |
1.00000 |
1
|
559
|