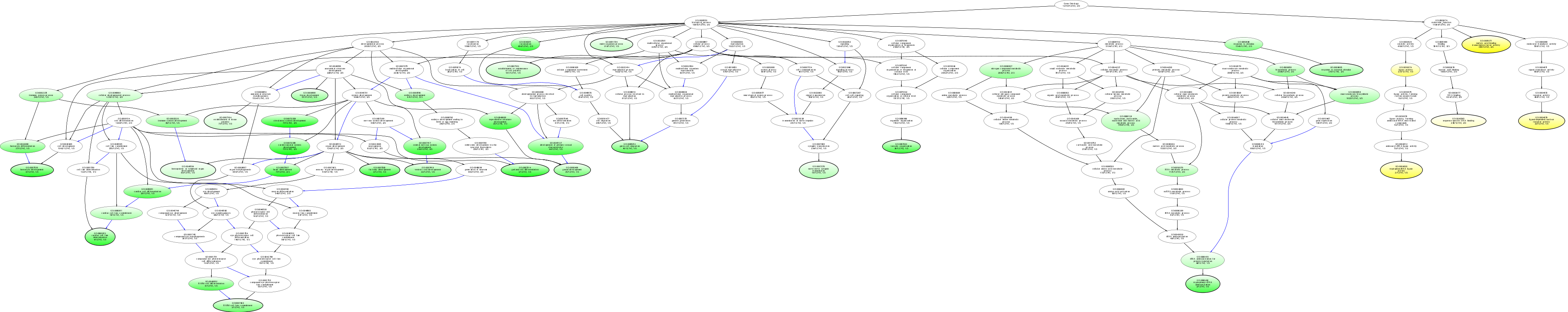
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0072359
|
circulatory system development
|
0.00203568 |
0.00203568 |
2
|
77
|
|
GO:0072358
|
cardiovascular system development
|
0.00203568 |
0.00203568 |
2
|
77
|
|
GO:0001071
|
nucleic acid binding transcription factor activity
|
0.00372409 |
0.00372409 |
2
|
395
|
|
GO:0007507
|
heart development
|
0.00379674 |
0.00379674 |
2
|
77
|
|
GO:0007514
|
garland cell differentiation
|
0.00439238 |
0.00439238 |
1
|
3
|
|
GO:0007503
|
fat body development
|
0.00482703 |
0.00482703 |
1
|
3
|
|
GO:0007516
|
hemocyte development
|
0.00570342 |
0.00570342 |
1
|
6
|
|
GO:0040011
|
locomotion
|
0.00603506 |
0.00603506 |
2
|
484
|
|
GO:0007417
|
central nervous system development
|
0.0183218 |
0.0183218 |
2
|
230
|
|
GO:0060913
|
cardiac cell fate determination
|
0.0285714 |
0.0285714 |
1
|
4
|
|
GO:0048608
|
reproductive structure development
|
0.0304066 |
0.0304066 |
1
|
35
|
|
GO:0007033
|
vacuole organization
|
0.0304569 |
0.0304569 |
1
|
36
|
|
GO:0045137
|
development of primary sexual characteristics
|
0.0351060 |
0.0351060 |
1
|
44
|
|
GO:0004830
|
tryptophan-tRNA ligase activity
|
0.0377358 |
0.0377358 |
1
|
2
|
|
GO:0006436
|
tryptophanyl-tRNA aminoacylation
|
0.0408163 |
0.0408163 |
1
|
2
|
|
GO:0035051
|
cardiac cell differentiation
|
0.0441064 |
0.0441064 |
1
|
28
|
|
GO:0048052
|
R1/R6 cell differentiation
|
0.0447761 |
0.0447761 |
1
|
6
|
|
GO:0008354
|
germ cell migration
|
0.0472279 |
0.0472279 |
1
|
46
|
|
GO:0004879
|
ligand-dependent nuclear receptor activity
|
0.0501089 |
0.0501089 |
1
|
23
|
|
GO:0007419
|
ventral cord development
|
0.0508390 |
0.0508390 |
1
|
33
|
|
GO:0042386
|
hemocyte differentiation
|
0.0522876 |
0.0522876 |
1
|
37
|
|
GO:0007462
|
R1/R6 cell fate commitment
|
0.0526316 |
0.0526316 |
1
|
3
|
|
GO:0008406
|
gonad development
|
0.0555886 |
0.0555886 |
1
|
35
|
|
GO:0016874
|
ligase activity
|
0.0562591 |
0.0562591 |
1
|
231
|
|
GO:0050896
|
response to stimulus
|
0.0575624 |
0.0575624 |
2
|
1548
|
|
GO:0009605
|
response to external stimulus
|
0.0585655 |
0.0585655 |
2
|
375
|
|
GO:0009790
|
embryo development
|
0.0589730 |
0.0589730 |
2
|
641
|
|
GO:0009888
|
tissue development
|
0.0603928 |
0.0603928 |
2
|
557
|
|
GO:0002520
|
immune system development
|
0.0644123 |
0.0644123 |
1
|
60
|
|
GO:0009058
|
biosynthetic process
|
0.0650969 |
0.0650969 |
3
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.0653778 |
0.0653778 |
3
|
2093
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.0666764 |
0.0666764 |
3
|
1623
|
|
GO:0002376
|
immune system process
|
0.0676558 |
0.0676558 |
1
|
245
|
|
GO:0060911
|
cardiac cell fate commitment
|
0.0687023 |
0.0687023 |
1
|
18
|
|
GO:0016070
|
RNA metabolic process
|
0.0707654 |
0.0707654 |
3
|
1191
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.0717945 |
0.0717945 |
3
|
1789
|
|
GO:0007163
|
establishment or maintenance of cell polarity
|
0.0718465 |
0.0718465 |
1
|
161
|
|
GO:0007270
|
nerve-nerve synaptic transmission
|
0.0769231 |
0.0769231 |
1
|
18
|
|
GO:0006418
|
tRNA aminoacylation for protein translation
|
0.0816667 |
0.0816667 |
1
|
49
|
|
GO:0048869
|
cellular developmental process
|
0.0826655 |
0.0826655 |
2
|
1276
|
|
GO:0051704
|
multi-organism process
|
0.0874732 |
0.0874732 |
1
|
319
|
|
GO:0043565
|
sequence-specific DNA binding
|
0.0903278 |
0.0903278 |
2
|
248
|
|
GO:0048534
|
hemopoietic or lymphoid organ development
|
0.0943216 |
0.0943216 |
1
|
60
|
|
GO:0007164
|
establishment of tissue polarity
|
0.0979026 |
0.0979026 |
1
|
77
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.102602 |
0.102602 |
2
|
2108
|
|
GO:0007548
|
sex differentiation
|
0.111296 |
0.111296 |
1
|
67
|
|
GO:0042067
|
establishment of ommatidial planar polarity
|
0.111814 |
0.111814 |
1
|
53
|
|
GO:0048056
|
R3/R4 cell differentiation
|
0.111940 |
0.111940 |
1
|
15
|
|
GO:0030528
|
transcription regulator activity
|
0.112632 |
0.112632 |
1
|
432
|
|
GO:0019222
|
regulation of metabolic process
|
0.114129 |
0.114129 |
2
|
1242
|
|
GO:0050789
|
regulation of biological process
|
0.115315 |
0.115315 |
2
|
2246
|
|
GO:0006520
|
cellular amino acid metabolic process
|
0.115315 |
0.115315 |
1
|
172
|
|
GO:0008152
|
metabolic process
|
0.117083 |
0.117083 |
3
|
5194
|
|
GO:0006928
|
cellular component movement
|
0.118060 |
0.118060 |
1
|
269
|
|
GO:0044237
|
cellular metabolic process
|
0.118315 |
0.118315 |
3
|
3814
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.122595 |
0.122595 |
3
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.123669 |
0.123669 |
3
|
1959
|
|
GO:0005488
|
binding
|
0.132501 |
0.132501 |
3
|
5641
|
|
GO:0051674
|
localization of cell
|
0.138713 |
0.138713 |
1
|
263
|
|
GO:0009792
|
embryo development ending in birth or egg hatching
|
0.139821 |
0.139821 |
2
|
240
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.142051 |
0.142051 |
2
|
1039
|
|
GO:0060089
|
molecular transducer activity
|
0.144056 |
0.144056 |
1
|
559
|
|
GO:0065007
|
biological regulation
|
0.145142 |
0.145142 |
2
|
2548
|
|
GO:0044249
|
cellular biosynthetic process
|
0.146100 |
0.146100 |
3
|
2034
|
|
GO:0048542
|
lymph gland development
|
0.146226 |
0.146226 |
1
|
31
|
|
GO:0010467
|
gene expression
|
0.150029 |
0.150029 |
3
|
1907
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.150277 |
0.150277 |
2
|
1110
|
|
GO:0042330
|
taxis
|
0.151554 |
0.151554 |
2
|
237
|
|
GO:0007464
|
R3/R4 cell fate commitment
|
0.153846 |
0.153846 |
1
|
14
|
|
GO:0032502
|
developmental process
|
0.154531 |
0.154531 |
2
|
2638
|
|
GO:0043167
|
ion binding
|
0.174057 |
0.174057 |
2
|
1498
|
|
GO:0048663
|
neuron fate commitment
|
0.179754 |
0.179754 |
1
|
64
|
|
GO:0007267
|
cell-cell signaling
|
0.188801 |
0.188801 |
1
|
263
|
|
GO:0006082
|
organic acid metabolic process
|
0.189003 |
0.189003 |
1
|
259
|
|
GO:0008045
|
motor axon guidance
|
0.193548 |
0.193548 |
1
|
36
|
|
GO:0019226
|
transmission of nerve impulse
|
0.194826 |
0.194826 |
1
|
241
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.194907 |
0.194907 |
2
|
1056
|
|
GO:0030182
|
neuron differentiation
|
0.195423 |
0.195423 |
2
|
548
|
|
GO:0009607
|
response to biotic stimulus
|
0.196095 |
0.196095 |
1
|
160
|
|
GO:0001709
|
cell fate determination
|
0.196268 |
0.196268 |
1
|
132
|
|
GO:0007154
|
cell communication
|
0.196383 |
0.196383 |
1
|
461
|
|
GO:0003677
|
DNA binding
|
0.197185 |
0.197185 |
2
|
824
|
|
GO:0023046
|
signaling process
|
0.199779 |
0.199779 |
1
|
760
|
|
GO:0009453
|
energy taxis
|
0.200243 |
0.200243 |
1
|
25
|
|
GO:0042180
|
cellular ketone metabolic process
|
0.203187 |
0.203187 |
1
|
280
|
|
GO:0032774
|
RNA biosynthetic process
|
0.203435 |
0.203435 |
2
|
703
|
|
GO:0022008
|
neurogenesis
|
0.204486 |
0.204486 |
2
|
632
|
|
GO:0030030
|
cell projection organization
|
0.205631 |
0.205631 |
1
|
485
|
|
GO:0061061
|
muscle structure development
|
0.210182 |
0.210182 |
1
|
252
|
|
GO:0006955
|
immune response
|
0.214169 |
0.214169 |
1
|
180
|
|
GO:0050794
|
regulation of cellular process
|
0.224039 |
0.224039 |
2
|
2034
|
|
GO:0042331
|
phototaxis
|
0.225225 |
0.225225 |
1
|
25
|
|
GO:0016875
|
ligase activity, forming carbon-oxygen bonds
|
0.229437 |
0.229437 |
1
|
53
|
|
GO:0032501
|
multicellular organismal process
|
0.231607 |
0.231607 |
2
|
3315
|
|
GO:0009987
|
cellular process
|
0.235914 |
0.235914 |
3
|
6560
|
|
GO:0048610
|
cellular process involved in reproduction
|
0.248418 |
0.248418 |
1
|
613
|
|
GO:0007399
|
nervous system development
|
0.249853 |
0.249853 |
2
|
848
|
|
GO:0003676
|
nucleic acid binding
|
0.253252 |
0.253252 |
2
|
1855
|
|
GO:0044106
|
cellular amine metabolic process
|
0.255042 |
0.255042 |
1
|
195
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.260978 |
0.260978 |
3
|
2878
|
|
GO:0000003
|
reproduction
|
0.261920 |
0.261920 |
1
|
1022
|
|
GO:0022414
|
reproductive process
|
0.261920 |
0.261920 |
1
|
1022
|
|
GO:0007510
|
cardioblast cell fate determination
|
0.266667 |
0.266667 |
1
|
4
|
|
GO:0034660
|
ncRNA metabolic process
|
0.271000 |
0.271000 |
1
|
119
|
|
GO:0007517
|
muscle organ development
|
0.276072 |
0.276072 |
1
|
191
|
|
GO:0048732
|
gland development
|
0.284024 |
0.284024 |
1
|
191
|
|
GO:0043038
|
amino acid activation
|
0.290698 |
0.290698 |
1
|
50
|
|
GO:0048513
|
organ development
|
0.300572 |
0.300572 |
2
|
1242
|
|
GO:0009628
|
response to abiotic stimulus
|
0.302411 |
0.302411 |
1
|
255
|
|
GO:0001738
|
morphogenesis of a polarized epithelium
|
0.304498 |
0.304498 |
1
|
88
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.304837 |
0.304837 |
2
|
888
|
|
GO:0007498
|
mesoderm development
|
0.312278 |
0.312278 |
1
|
95
|
|
GO:0045466
|
R7 cell differentiation
|
0.313433 |
0.313433 |
1
|
42
|
|
GO:0007465
|
R7 cell fate commitment
|
0.323529 |
0.323529 |
1
|
22
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.325784 |
0.325784 |
2
|
905
|
|
GO:0043170
|
macromolecule metabolic process
|
0.329564 |
0.329564 |
3
|
3588
|
|
GO:0005575
|
cellular_component
|
0.330897 |
0.330897 |
3
|
8817
|
|
GO:0006350
|
transcription
|
0.334468 |
0.334468 |
2
|
859
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.338051 |
0.338051 |
2
|
1534
|
|
GO:0023052
|
signaling
|
0.338079 |
0.338079 |
1
|
1364
|
|
GO:0046552
|
photoreceptor cell fate commitment
|
0.345238 |
0.345238 |
1
|
58
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.346544 |
0.346544 |
2
|
888
|
|
GO:0005623
|
cell
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0044464
|
cell part
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0048729
|
tissue morphogenesis
|
0.350704 |
0.350704 |
1
|
312
|
|
GO:0043226
|
organelle
|
0.353651 |
0.353651 |
2
|
3537
|
|
GO:0044424
|
intracellular part
|
0.354324 |
0.354324 |
3
|
4384
|
|
GO:0001751
|
compound eye photoreceptor cell differentiation
|
0.354497 |
0.354497 |
1
|
134
|
|
GO:0045165
|
cell fate commitment
|
0.359875 |
0.359875 |
1
|
255
|
|
GO:0003008
|
system process
|
0.360531 |
0.360531 |
1
|
664
|
|
GO:0001754
|
eye photoreceptor cell differentiation
|
0.361386 |
0.361386 |
1
|
146
|
|
GO:0001882
|
nucleoside binding
|
0.361738 |
0.361738 |
1
|
784
|
|
GO:0003006
|
developmental process involved in reproduction
|
0.367874 |
0.367874 |
1
|
602
|
|
GO:0042706
|
eye photoreceptor cell fate commitment
|
0.371622 |
0.371622 |
1
|
55
|
|
GO:0000904
|
cell morphogenesis involved in differentiation
|
0.376731 |
0.376731 |
1
|
408
|
|
GO:0044281
|
small molecule metabolic process
|
0.378352 |
0.378352 |
1
|
761
|
|
GO:0009308
|
amine metabolic process
|
0.397406 |
0.397406 |
1
|
325
|
|
GO:0005634
|
nucleus
|
0.408695 |
0.408695 |
2
|
1826
|
|
GO:0001752
|
compound eye photoreceptor fate commitment
|
0.410448 |
0.410448 |
1
|
55
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.410831 |
0.410831 |
2
|
686
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.427778 |
0.427778 |
2
|
903
|
|
GO:0048731
|
system development
|
0.432376 |
0.432376 |
2
|
1696
|
|
GO:0007280
|
pole cell migration
|
0.434783 |
0.434783 |
1
|
20
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.435609 |
0.435609 |
2
|
850
|
|
GO:0007275
|
multicellular organismal development
|
0.442066 |
0.442066 |
2
|
2296
|
|
GO:0051179
|
localization
|
0.445834 |
0.445834 |
1
|
1896
|
|
GO:0005622
|
intracellular
|
0.446165 |
0.446165 |
3
|
4734
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.468064 |
0.468064 |
2
|
620
|
|
GO:0045449
|
regulation of transcription
|
0.472005 |
0.472005 |
2
|
771
|
|
GO:0051707
|
response to other organism
|
0.473846 |
0.473846 |
1
|
154
|
|
GO:0032989
|
cellular component morphogenesis
|
0.474782 |
0.474782 |
1
|
566
|
|
GO:0048609
|
multicellular organismal reproductive process
|
0.480389 |
0.480389 |
1
|
937
|
|
GO:0032504
|
multicellular organism reproduction
|
0.480389 |
0.480389 |
1
|
937
|
|
GO:0010468
|
regulation of gene expression
|
0.484889 |
0.484889 |
2
|
973
|
|
GO:0009314
|
response to radiation
|
0.490196 |
0.490196 |
1
|
125
|
|
GO:0006935
|
chemotaxis
|
0.492835 |
0.492835 |
1
|
193
|
|
GO:0046530
|
photoreceptor cell differentiation
|
0.504230 |
0.504230 |
1
|
162
|
|
GO:0000166
|
nucleotide binding
|
0.504833 |
0.504833 |
1
|
1178
|
|
GO:0006412
|
translation
|
0.514942 |
0.514942 |
1
|
600
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.520400 |
0.520400 |
2
|
850
|
|
GO:0048592
|
eye morphogenesis
|
0.542069 |
0.542069 |
1
|
393
|
|
GO:0044238
|
primary metabolic process
|
0.551267 |
0.551267 |
3
|
4259
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.556542 |
0.556542 |
1
|
1562
|
|
GO:0005737
|
cytoplasm
|
0.563502 |
0.563502 |
2
|
2378
|
|
GO:0043039
|
tRNA aminoacylation
|
0.576471 |
0.576471 |
1
|
49
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0019730
|
antimicrobial humoral response
|
0.584337 |
0.584337 |
1
|
97
|
|
GO:0009887
|
organ morphogenesis
|
0.608733 |
0.608733 |
1
|
684
|
|
GO:0008270
|
zinc ion binding
|
0.610500 |
0.610500 |
2
|
901
|
|
GO:0006959
|
humoral immune response
|
0.616667 |
0.616667 |
1
|
111
|
|
GO:0003702
|
RNA polymerase II transcription factor activity
|
0.618056 |
0.618056 |
1
|
267
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.631500 |
0.631500 |
3
|
1535
|
|
GO:0046914
|
transition metal ion binding
|
0.633060 |
0.633060 |
2
|
1153
|
|
GO:0042221
|
response to chemical stimulus
|
0.650010 |
0.650010 |
1
|
632
|
|
GO:0043227
|
membrane-bounded organelle
|
0.653325 |
0.653325 |
2
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.653801 |
0.653801 |
2
|
2856
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.662696 |
0.662696 |
2
|
701
|
|
GO:0005515
|
protein binding
|
0.665786 |
0.665786 |
1
|
1726
|
|
GO:0007409
|
axonogenesis
|
0.679389 |
0.679389 |
1
|
267
|
|
GO:0048666
|
neuron development
|
0.679533 |
0.679533 |
1
|
471
|
|
GO:0007411
|
axon guidance
|
0.683824 |
0.683824 |
1
|
186
|
|
GO:0007423
|
sensory organ development
|
0.684358 |
0.684358 |
1
|
544
|
|
GO:0031175
|
neuron projection development
|
0.696270 |
0.696270 |
1
|
392
|
|
GO:0048468
|
cell development
|
0.705715 |
0.705715 |
1
|
1042
|
|
GO:0006399
|
tRNA metabolic process
|
0.705882 |
0.705882 |
1
|
84
|
|
GO:0010002
|
cardioblast differentiation
|
0.714286 |
0.714286 |
1
|
20
|
|
GO:0042684
|
cardioblast cell fate commitment
|
0.714286 |
0.714286 |
1
|
15
|
|
GO:0048856
|
anatomical structure development
|
0.737157 |
0.737157 |
2
|
2265
|
|
GO:0032990
|
cell part morphogenesis
|
0.745583 |
0.745583 |
1
|
422
|
|
GO:0032553
|
ribonucleotide binding
|
0.746180 |
0.746180 |
1
|
879
|
|
GO:0003824
|
catalytic activity
|
0.751316 |
0.751316 |
1
|
4106
|
|
GO:0032559
|
adenyl ribonucleotide binding
|
0.757705 |
0.757705 |
1
|
713
|
|
GO:0048870
|
cell motility
|
0.762455 |
0.762455 |
1
|
254
|
|
GO:0006996
|
organelle organization
|
0.779169 |
0.779169 |
1
|
1182
|
|
GO:0060429
|
epithelium development
|
0.779194 |
0.779194 |
1
|
295
|
|
GO:0048667
|
cell morphogenesis involved in neuron differentiation
|
0.795501 |
0.795501 |
1
|
389
|
|
GO:0017076
|
purine nucleotide binding
|
0.803056 |
0.803056 |
1
|
946
|
|
GO:0048858
|
cell projection morphogenesis
|
0.806883 |
0.806883 |
1
|
422
|
|
GO:0001745
|
compound eye morphogenesis
|
0.811947 |
0.811947 |
1
|
367
|
|
GO:0030554
|
adenyl nucleotide binding
|
0.819239 |
0.819239 |
1
|
775
|
|
GO:0004872
|
receptor activity
|
0.821109 |
0.821109 |
1
|
459
|
|
GO:0044267
|
cellular protein metabolic process
|
0.824560 |
0.824560 |
1
|
1505
|
|
GO:0000902
|
cell morphogenesis
|
0.825088 |
0.825088 |
1
|
467
|
|
GO:0001654
|
eye development
|
0.841912 |
0.841912 |
1
|
458
|
|
GO:0001736
|
establishment of planar polarity
|
0.853933 |
0.853933 |
1
|
76
|
|
GO:0019538
|
protein metabolic process
|
0.855761 |
0.855761 |
1
|
2053
|
|
GO:0019953
|
sexual reproduction
|
0.859100 |
0.859100 |
1
|
878
|
|
GO:0007268
|
synaptic transmission
|
0.866667 |
0.866667 |
1
|
234
|
|
GO:0007276
|
gamete generation
|
0.883697 |
0.883697 |
1
|
851
|
|
GO:0009416
|
response to light stimulus
|
0.888000 |
0.888000 |
1
|
111
|
|
GO:0043229
|
intracellular organelle
|
0.899971 |
0.899971 |
2
|
3530
|
|
GO:0002009
|
morphogenesis of an epithelium
|
0.908805 |
0.908805 |
1
|
289
|
|
GO:0048699
|
generation of neurons
|
0.910264 |
0.910264 |
2
|
603
|
|
GO:0048812
|
neuron projection morphogenesis
|
0.910798 |
0.910798 |
1
|
388
|
|
GO:0003707
|
steroid hormone receptor activity
|
0.913043 |
0.913043 |
1
|
21
|
|
GO:0007040
|
lysosome organization
|
0.916667 |
0.916667 |
1
|
33
|
|
GO:0001700
|
embryonic development via the syncytial blastoderm
|
0.918236 |
0.918236 |
2
|
230
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.923977 |
0.923977 |
1
|
1517
|
|
GO:0043436
|
oxoacid metabolic process
|
0.925000 |
0.925000 |
1
|
259
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.929175 |
0.929175 |
1
|
879
|
|
GO:0048749
|
compound eye development
|
0.932314 |
0.932314 |
1
|
427
|
|
GO:0046872
|
metal ion binding
|
0.936879 |
0.936879 |
2
|
1449
|
|
GO:0030154
|
cell differentiation
|
0.942825 |
0.942825 |
2
|
1239
|
|
GO:0016043
|
cellular component organization
|
0.947563 |
0.947563 |
2
|
2052
|
|
GO:0050877
|
neurological system process
|
0.953313 |
0.953313 |
1
|
633
|
|
GO:0016477
|
cell migration
|
0.964567 |
0.964567 |
1
|
245
|
|
GO:0001883
|
purine nucleoside binding
|
0.992347 |
0.992347 |
1
|
778
|
|
GO:0005524
|
ATP binding
|
0.998597 |
0.998597 |
1
|
712
|
|
GO:0043169
|
cation binding
|
0.998665 |
0.998665 |
2
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0016876
|
ligase activity, forming aminoacyl-tRNA and related compounds
|
1.00000 |
1.00000 |
1
|
53
|
|
GO:0019752
|
carboxylic acid metabolic process
|
1.00000 |
1.00000 |
1
|
259
|
|
GO:0003700
|
sequence-specific DNA binding transcription factor activity
|
1.00000 |
1.00000 |
2
|
395
|
|
GO:0004871
|
signal transducer activity
|
1.00000 |
1.00000 |
1
|
559
|
|
GO:0004812
|
aminoacyl-tRNA ligase activity
|
1.00000 |
1.00000 |
1
|
53
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|