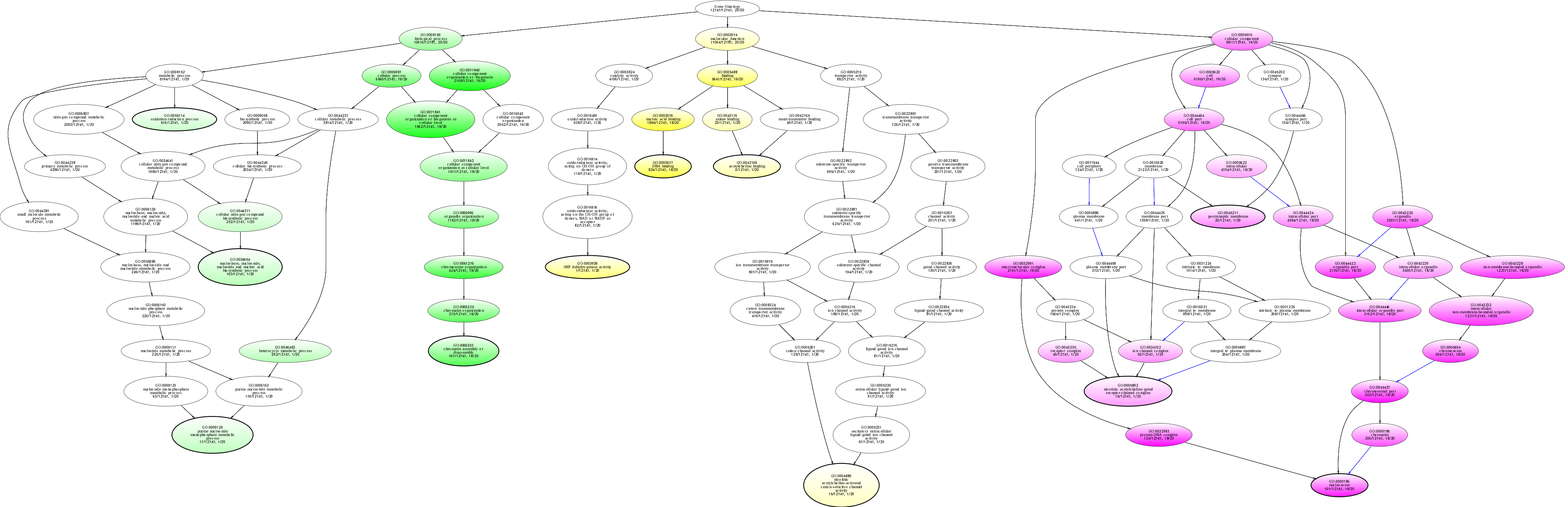
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0032993
|
protein-DNA complex
|
2.62926e-22 |
2.62926e-22 |
18
|
124
|
|
GO:0044427
|
chromosomal part
|
2.09311e-15 |
2.09311e-15 |
18
|
342
|
|
GO:0071840
|
cellular component organization or biogenesis
|
6.96505e-13 |
6.96505e-13 |
19
|
2108
|
|
GO:0032991
|
macromolecular complex
|
2.15217e-12 |
2.15217e-12 |
19
|
2151
|
|
GO:0000786
|
nucleosome
|
5.99915e-11 |
5.99915e-11 |
18
|
101
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
7.84919e-11 |
7.84919e-11 |
18
|
1562
|
|
GO:0044422
|
organelle part
|
1.59561e-10 |
1.59561e-10 |
18
|
2176
|
|
GO:0005694
|
chromosome
|
1.00680e-09 |
1.00680e-09 |
18
|
394
|
|
GO:0043228
|
non-membrane-bounded organelle
|
4.87725e-09 |
4.87725e-09 |
18
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
5.05474e-09 |
5.05474e-09 |
18
|
1227
|
|
GO:0051276
|
chromosome organization
|
7.63923e-09 |
7.63923e-09 |
18
|
424
|
|
GO:0003676
|
nucleic acid binding
|
2.50932e-08 |
2.50932e-08 |
18
|
1855
|
|
GO:0003677
|
DNA binding
|
4.08523e-07 |
4.08523e-07 |
18
|
824
|
|
GO:0043226
|
organelle
|
8.32352e-07 |
8.32352e-07 |
18
|
3537
|
|
GO:0044446
|
intracellular organelle part
|
2.85960e-06 |
2.85960e-06 |
18
|
2162
|
|
GO:0005488
|
binding
|
2.81205e-05 |
2.81205e-05 |
19
|
5641
|
|
GO:0000785
|
chromatin
|
4.58552e-05 |
4.58552e-05 |
18
|
200
|
|
GO:0006333
|
chromatin assembly or disassembly
|
9.86682e-05 |
9.86682e-05 |
18
|
167
|
|
GO:0006325
|
chromatin organization
|
0.000294096 |
0.000294096 |
18
|
273
|
|
GO:0009987
|
cellular process
|
0.000873388 |
0.000873388 |
19
|
6560
|
|
GO:0005623
|
cell
|
0.00121372 |
0.00121372 |
19
|
6195
|
|
GO:0044464
|
cell part
|
0.00121372 |
0.00121372 |
19
|
6195
|
|
GO:0005575
|
cellular_component
|
0.00620134 |
0.00620134 |
19
|
8817
|
|
GO:0006996
|
organelle organization
|
0.0108855 |
0.0108855 |
18
|
1182
|
|
GO:0003938
|
IMP dehydrogenase activity
|
0.0121951 |
0.0121951 |
1
|
1
|
|
GO:0044424
|
intracellular part
|
0.0123100 |
0.0123100 |
18
|
4384
|
|
GO:0045211
|
postsynaptic membrane
|
0.0139211 |
0.0139211 |
1
|
30
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.0175015 |
0.0175015 |
18
|
1517
|
|
GO:0043229
|
intracellular organelle
|
0.0195833 |
0.0195833 |
18
|
3530
|
|
GO:0034702
|
ion channel complex
|
0.0210151 |
0.0210151 |
1
|
53
|
|
GO:0043235
|
receptor complex
|
0.0255754 |
0.0255754 |
1
|
40
|
|
GO:0008150
|
biological_process
|
0.0256896 |
0.0256896 |
20
|
10616
|
|
GO:0005892
|
nicotinic acetylcholine-gated receptor-channel complex
|
0.0391645 |
0.0391645 |
1
|
15
|
|
GO:0005622
|
intracellular
|
0.0411926 |
0.0411926 |
18
|
4734
|
|
GO:0042166
|
acetylcholine binding
|
0.0434783 |
0.0434783 |
1
|
3
|
|
GO:0003674
|
molecular_function
|
0.0587622 |
0.0587622 |
20
|
11064
|
|
GO:0043176
|
amine binding
|
0.0748055 |
0.0748055 |
1
|
23
|
|
GO:0046483
|
heterocycle metabolic process
|
0.0765600 |
0.0765600 |
1
|
292
|
|
GO:0034654
|
nucleobase, nucleoside, nucleotide and nucleic acid biosynthetic process
|
0.0870726 |
0.0870726 |
1
|
163
|
|
GO:0009126
|
purine nucleoside monophosphate metabolic process
|
0.0876289 |
0.0876289 |
1
|
17
|
|
GO:0004889
|
nicotinic acetylcholine-activated cation-selective channel activity
|
0.0882353 |
0.0882353 |
1
|
15
|
|
GO:0044271
|
cellular nitrogen compound biosynthetic process
|
0.0931953 |
0.0931953 |
1
|
252
|
|
GO:0055114
|
oxidation-reduction process
|
0.0993454 |
0.0993454 |
1
|
516
|
|
GO:0044283
|
small molecule biosynthetic process
|
0.105199 |
0.105199 |
1
|
259
|
|
GO:0055086
|
nucleobase, nucleoside and nucleotide metabolic process
|
0.112568 |
0.112568 |
1
|
249
|
|
GO:0009167
|
purine ribonucleoside monophosphate metabolic process
|
0.114094 |
0.114094 |
1
|
17
|
|
GO:0009161
|
ribonucleoside monophosphate metabolic process
|
0.115183 |
0.115183 |
1
|
22
|
|
GO:0009127
|
purine nucleoside monophosphate biosynthetic process
|
0.115646 |
0.115646 |
1
|
17
|
|
GO:0006177
|
GMP biosynthetic process
|
0.117647 |
0.117647 |
1
|
2
|
|
GO:0046037
|
GMP metabolic process
|
0.117647 |
0.117647 |
1
|
2
|
|
GO:0005811
|
lipid particle
|
0.134192 |
0.134192 |
1
|
250
|
|
GO:0044281
|
small molecule metabolic process
|
0.146515 |
0.146515 |
1
|
761
|
|
GO:0009156
|
ribonucleoside monophosphate biosynthetic process
|
0.150685 |
0.150685 |
1
|
22
|
|
GO:0042165
|
neurotransmitter binding
|
0.152978 |
0.152978 |
1
|
49
|
|
GO:0016491
|
oxidoreductase activity
|
0.155382 |
0.155382 |
1
|
638
|
|
GO:0009168
|
purine ribonucleoside monophosphate biosynthetic process
|
0.158879 |
0.158879 |
1
|
17
|
|
GO:0031226
|
intrinsic to plasma membrane
|
0.179931 |
0.179931 |
1
|
208
|
|
GO:0016614
|
oxidoreductase activity, acting on CH-OH group of donors
|
0.184953 |
0.184953 |
1
|
118
|
|
GO:0071824
|
protein-DNA complex subunit organization
|
0.195833 |
0.195833 |
1
|
47
|
|
GO:0071103
|
DNA conformation change
|
0.201202 |
0.201202 |
1
|
77
|
|
GO:0044456
|
synapse part
|
0.202024 |
0.202024 |
1
|
104
|
|
GO:0005887
|
integral to plasma membrane
|
0.203593 |
0.203593 |
1
|
204
|
|
GO:0044459
|
plasma membrane part
|
0.236675 |
0.236675 |
1
|
373
|
|
GO:0009605
|
response to external stimulus
|
0.242248 |
0.242248 |
1
|
375
|
|
GO:0045202
|
synapse
|
0.252691 |
0.252691 |
1
|
134
|
|
GO:0065004
|
protein-DNA complex assembly
|
0.267045 |
0.267045 |
1
|
47
|
|
GO:0005261
|
cation channel activity
|
0.272340 |
0.272340 |
1
|
128
|
|
GO:0022803
|
passive transmembrane transporter activity
|
0.276860 |
0.276860 |
1
|
201
|
|
GO:0009123
|
nucleoside monophosphate metabolic process
|
0.278761 |
0.278761 |
1
|
63
|
|
GO:0048609
|
multicellular organismal reproductive process
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0032504
|
multicellular organism reproduction
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0034622
|
cellular macromolecular complex assembly
|
0.286645 |
0.286645 |
1
|
176
|
|
GO:0006935
|
chemotaxis
|
0.287630 |
0.287630 |
1
|
193
|
|
GO:0005886
|
plasma membrane
|
0.292027 |
0.292027 |
1
|
641
|
|
GO:0065003
|
macromolecular complex assembly
|
0.301303 |
0.301303 |
1
|
185
|
|
GO:0022838
|
substrate-specific channel activity
|
0.307448 |
0.307448 |
1
|
194
|
|
GO:0005216
|
ion channel activity
|
0.369141 |
0.369141 |
1
|
189
|
|
GO:0000904
|
cell morphogenesis involved in differentiation
|
0.376731 |
0.376731 |
1
|
408
|
|
GO:0009124
|
nucleoside monophosphate biosynthetic process
|
0.384615 |
0.384615 |
1
|
60
|
|
GO:0042330
|
taxis
|
0.389803 |
0.389803 |
1
|
237
|
|
GO:0006811
|
ion transport
|
0.393444 |
0.393444 |
1
|
331
|
|
GO:0009058
|
biosynthetic process
|
0.402387 |
0.402387 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.402965 |
0.402965 |
1
|
2093
|
|
GO:0042221
|
response to chemical stimulus
|
0.408269 |
0.408269 |
1
|
632
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.415756 |
0.415756 |
1
|
1789
|
|
GO:0015276
|
ligand-gated ion channel activity
|
0.428571 |
0.428571 |
1
|
81
|
|
GO:0048666
|
neuron development
|
0.433702 |
0.433702 |
1
|
471
|
|
GO:0030182
|
neuron differentiation
|
0.442292 |
0.442292 |
1
|
548
|
|
GO:0006334
|
nucleosome assembly
|
0.442623 |
0.442623 |
1
|
27
|
|
GO:0022008
|
neurogenesis
|
0.452398 |
0.452398 |
1
|
632
|
|
GO:0048468
|
cell development
|
0.457419 |
0.457419 |
1
|
1042
|
|
GO:0034404
|
nucleobase, nucleoside and nucleotide biosynthetic process
|
0.473837 |
0.473837 |
1
|
163
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.498346 |
0.498346 |
1
|
1959
|
|
GO:0007399
|
nervous system development
|
0.500000 |
0.500000 |
1
|
848
|
|
GO:0044249
|
cellular biosynthetic process
|
0.526807 |
0.526807 |
1
|
2034
|
|
GO:0006163
|
purine nucleotide metabolic process
|
0.528529 |
0.528529 |
1
|
176
|
|
GO:0009260
|
ribonucleotide biosynthetic process
|
0.553846 |
0.553846 |
1
|
108
|
|
GO:0009152
|
purine ribonucleotide biosynthetic process
|
0.569832 |
0.569832 |
1
|
102
|
|
GO:0006323
|
DNA packaging
|
0.573849 |
0.573849 |
1
|
70
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0031497
|
chromatin assembly
|
0.583715 |
0.583715 |
1
|
34
|
|
GO:0016043
|
cellular component organization
|
0.598220 |
0.598220 |
19
|
2052
|
|
GO:0040011
|
locomotion
|
0.607071 |
0.607071 |
1
|
484
|
|
GO:0022834
|
ligand-gated channel activity
|
0.623077 |
0.623077 |
1
|
81
|
|
GO:0016044
|
cellular membrane organization
|
0.641154 |
0.641154 |
1
|
344
|
|
GO:0022836
|
gated channel activity
|
0.646766 |
0.646766 |
1
|
130
|
|
GO:0009165
|
nucleotide biosynthetic process
|
0.648305 |
0.648305 |
1
|
153
|
|
GO:0006897
|
endocytosis
|
0.655814 |
0.655814 |
1
|
282
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0006164
|
purine nucleotide biosynthetic process
|
0.661616 |
0.661616 |
1
|
131
|
|
GO:0009259
|
ribonucleotide metabolic process
|
0.663717 |
0.663717 |
1
|
150
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0007409
|
axonogenesis
|
0.679389 |
0.679389 |
1
|
267
|
|
GO:0006911
|
phagocytosis, engulfment
|
0.680851 |
0.680851 |
1
|
192
|
|
GO:0007411
|
axon guidance
|
0.683824 |
0.683824 |
1
|
186
|
|
GO:0016616
|
oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor
|
0.694915 |
0.694915 |
1
|
82
|
|
GO:0031175
|
neuron projection development
|
0.696270 |
0.696270 |
1
|
392
|
|
GO:0006909
|
phagocytosis
|
0.723404 |
0.723404 |
1
|
204
|
|
GO:0031224
|
intrinsic to membrane
|
0.725322 |
0.725322 |
1
|
1014
|
|
GO:0016192
|
vesicle-mediated transport
|
0.726944 |
0.726944 |
1
|
430
|
|
GO:0007292
|
female gamete generation
|
0.727380 |
0.727380 |
1
|
619
|
|
GO:0032990
|
cell part morphogenesis
|
0.745583 |
0.745583 |
1
|
422
|
|
GO:0005230
|
extracellular ligand-gated ion channel activity
|
0.753086 |
0.753086 |
1
|
61
|
|
GO:0030030
|
cell projection organization
|
0.767724 |
0.767724 |
1
|
485
|
|
GO:0034728
|
nucleosome organization
|
0.772860 |
0.772860 |
1
|
36
|
|
GO:0022891
|
substrate-specific transmembrane transporter activity
|
0.788146 |
0.788146 |
1
|
625
|
|
GO:0009150
|
purine ribonucleotide metabolic process
|
0.791209 |
0.791209 |
1
|
144
|
|
GO:0048667
|
cell morphogenesis involved in neuron differentiation
|
0.795501 |
0.795501 |
1
|
389
|
|
GO:0005215
|
transporter activity
|
0.798929 |
0.798929 |
1
|
852
|
|
GO:0048858
|
cell projection morphogenesis
|
0.806883 |
0.806883 |
1
|
422
|
|
GO:0008324
|
cation transmembrane transporter activity
|
0.808679 |
0.808679 |
1
|
410
|
|
GO:0015075
|
ion transmembrane transporter activity
|
0.811200 |
0.811200 |
1
|
507
|
|
GO:0051234
|
establishment of localization
|
0.811836 |
0.811836 |
2
|
1549
|
|
GO:0022892
|
substrate-specific transporter activity
|
0.815728 |
0.815728 |
1
|
695
|
|
GO:0010324
|
membrane invagination
|
0.819767 |
0.819767 |
1
|
282
|
|
GO:0044238
|
primary metabolic process
|
0.819985 |
0.819985 |
1
|
4259
|
|
GO:0000902
|
cell morphogenesis
|
0.825088 |
0.825088 |
1
|
467
|
|
GO:0022857
|
transmembrane transporter activity
|
0.852113 |
0.852113 |
1
|
726
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0019953
|
sexual reproduction
|
0.859100 |
0.859100 |
1
|
878
|
|
GO:0000003
|
reproduction
|
0.868190 |
0.868190 |
1
|
1022
|
|
GO:0022414
|
reproductive process
|
0.868190 |
0.868190 |
1
|
1022
|
|
GO:0007276
|
gamete generation
|
0.883697 |
0.883697 |
1
|
851
|
|
GO:0051179
|
localization
|
0.895665 |
0.895665 |
2
|
1896
|
|
GO:0071944
|
cell periphery
|
0.906051 |
0.906051 |
1
|
724
|
|
GO:0043933
|
macromolecular complex subunit organization
|
0.906930 |
0.906930 |
1
|
240
|
|
GO:0006753
|
nucleoside phosphate metabolic process
|
0.907631 |
0.907631 |
1
|
226
|
|
GO:0048812
|
neuron projection morphogenesis
|
0.910798 |
0.910798 |
1
|
388
|
|
GO:0006810
|
transport
|
0.933966 |
0.933966 |
2
|
1497
|
|
GO:0005231
|
excitatory extracellular ligand-gated ion channel activity
|
0.934426 |
0.934426 |
1
|
57
|
|
GO:0034621
|
cellular macromolecular complex subunit organization
|
0.949970 |
0.949970 |
1
|
232
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0050896
|
response to stimulus
|
0.957373 |
0.957373 |
1
|
1548
|
|
GO:0061024
|
membrane organization
|
0.970235 |
0.970235 |
1
|
345
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0048869
|
cellular developmental process
|
0.975306 |
0.975306 |
1
|
1276
|
|
GO:0032989
|
cellular component morphogenesis
|
0.983274 |
0.983274 |
1
|
566
|
|
GO:0016021
|
integral to membrane
|
0.984221 |
0.984221 |
1
|
998
|
|
GO:0048477
|
oogenesis
|
0.987076 |
0.987076 |
1
|
611
|
|
GO:0044425
|
membrane part
|
0.992306 |
0.992306 |
1
|
1398
|
|
GO:0032502
|
developmental process
|
0.996718 |
0.996718 |
1
|
2638
|
|
GO:0022607
|
cellular component assembly
|
0.997871 |
0.997871 |
1
|
577
|
|
GO:0044085
|
cellular component biogenesis
|
0.998894 |
0.998894 |
1
|
632
|
|
GO:0032501
|
multicellular organismal process
|
0.999444 |
0.999444 |
1
|
3315
|
|
GO:0016020
|
membrane
|
0.999659 |
0.999659 |
1
|
2122
|
|
GO:0071844
|
cellular component assembly at cellular level
|
0.999790 |
0.999790 |
1
|
568
|
|
GO:0003824
|
catalytic activity
|
0.999907 |
0.999907 |
1
|
4106
|
|
GO:0044444
|
cytoplasmic part
|
0.999954 |
0.999954 |
1
|
1863
|
|
GO:0044237
|
cellular metabolic process
|
0.999997 |
0.999997 |
1
|
3814
|
|
GO:0008152
|
metabolic process
|
0.999999 |
0.999999 |
1
|
5194
|
|
GO:0005737
|
cytoplasm
|
0.999999 |
0.999999 |
1
|
2378
|
|
GO:0043234
|
protein complex
|
1.00000 |
1.00000 |
1
|
1564
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
20
|
12747
|
|
GO:0009117
|
nucleotide metabolic process
|
1.00000 |
1.00000 |
1
|
226
|
|
GO:0015267
|
channel activity
|
1.00000 |
1.00000 |
1
|
201
|