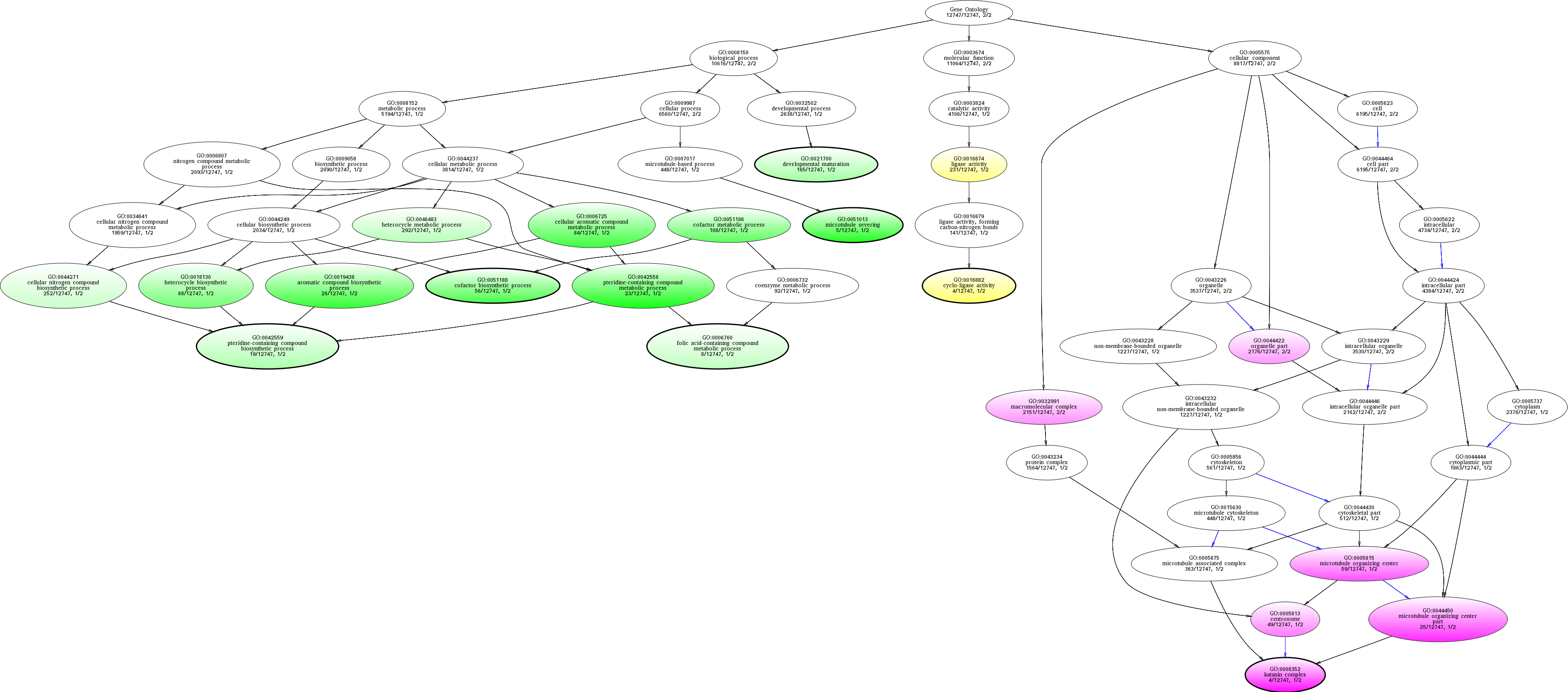
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0008352
|
katanin complex
|
0.0100503 |
0.0100503 |
1
|
4
|
|
GO:0042558
|
pteridine-containing compound metabolic process
|
0.0108902 |
0.0108902 |
1
|
23
|
|
GO:0051013
|
microtubule severing
|
0.0111607 |
0.0111607 |
1
|
5
|
|
GO:0044450
|
microtubule organizing center part
|
0.0116932 |
0.0116932 |
1
|
25
|
|
GO:0019438
|
aromatic compound biosynthetic process
|
0.0126521 |
0.0126521 |
1
|
26
|
|
GO:0006725
|
cellular aromatic compound metabolic process
|
0.0220241 |
0.0220241 |
1
|
84
|
|
GO:0051188
|
cofactor biosynthetic process
|
0.0268972 |
0.0268972 |
1
|
56
|
|
GO:0005815
|
microtubule organizing center
|
0.0275572 |
0.0275572 |
1
|
59
|
|
GO:0051186
|
cofactor metabolic process
|
0.0283167 |
0.0283167 |
1
|
108
|
|
GO:0016882
|
cyclo-ligase activity
|
0.0283688 |
0.0283688 |
1
|
4
|
|
GO:0018130
|
heterocycle biosynthetic process
|
0.0323040 |
0.0323040 |
1
|
68
|
|
GO:0005813
|
centrosome
|
0.0399348 |
0.0399348 |
1
|
49
|
|
GO:0016874
|
ligase activity
|
0.0562591 |
0.0562591 |
1
|
231
|
|
GO:0032991
|
macromolecular complex
|
0.0594958 |
0.0594958 |
2
|
2151
|
|
GO:0044422
|
organelle part
|
0.0608872 |
0.0608872 |
2
|
2176
|
|
GO:0021700
|
developmental maturation
|
0.0625474 |
0.0625474 |
1
|
165
|
|
GO:0042559
|
pteridine-containing compound biosynthetic process
|
0.0698529 |
0.0698529 |
1
|
19
|
|
GO:0046483
|
heterocycle metabolic process
|
0.0765600 |
0.0765600 |
1
|
292
|
|
GO:0006760
|
folic acid-containing compound metabolic process
|
0.0824742 |
0.0824742 |
1
|
8
|
|
GO:0044271
|
cellular nitrogen compound biosynthetic process
|
0.0931953 |
0.0931953 |
1
|
252
|
|
GO:0009994
|
oocyte differentiation
|
0.108434 |
0.108434 |
1
|
162
|
|
GO:0048469
|
cell maturation
|
0.121094 |
0.121094 |
1
|
155
|
|
GO:0008092
|
cytoskeletal protein binding
|
0.127462 |
0.127462 |
1
|
220
|
|
GO:0009396
|
folic acid-containing compound biosynthetic process
|
0.130435 |
0.130435 |
1
|
6
|
|
GO:0007017
|
microtubule-based process
|
0.131931 |
0.131931 |
1
|
448
|
|
GO:0043226
|
organelle
|
0.160900 |
0.160900 |
2
|
3537
|
|
GO:0005681
|
spliceosomal complex
|
0.167598 |
0.167598 |
1
|
180
|
|
GO:0048610
|
cellular process involved in reproduction
|
0.173346 |
0.173346 |
1
|
613
|
|
GO:0000003
|
reproduction
|
0.183280 |
0.183280 |
1
|
1022
|
|
GO:0022414
|
reproductive process
|
0.183280 |
0.183280 |
1
|
1022
|
|
GO:0007389
|
pattern specification process
|
0.203563 |
0.203563 |
1
|
537
|
|
GO:0003006
|
developmental process involved in reproduction
|
0.204901 |
0.204901 |
1
|
602
|
|
GO:0007309
|
oocyte axis specification
|
0.206278 |
0.206278 |
1
|
138
|
|
GO:0007308
|
oocyte construction
|
0.206997 |
0.206997 |
1
|
142
|
|
GO:0030529
|
ribonucleoprotein complex
|
0.210589 |
0.210589 |
1
|
510
|
|
GO:0006396
|
RNA processing
|
0.214286 |
0.214286 |
1
|
414
|
|
GO:0007281
|
germ cell development
|
0.219828 |
0.219828 |
1
|
306
|
|
GO:0005875
|
microtubule associated complex
|
0.224768 |
0.224768 |
1
|
363
|
|
GO:0044446
|
intracellular organelle part
|
0.243036 |
0.243036 |
2
|
2162
|
|
GO:0001882
|
nucleoside binding
|
0.258670 |
0.258670 |
1
|
784
|
|
GO:0005488
|
binding
|
0.259926 |
0.259926 |
2
|
5641
|
|
GO:0007310
|
oocyte dorsal/ventral axis specification
|
0.266272 |
0.266272 |
1
|
45
|
|
GO:0009950
|
dorsal/ventral axis specification
|
0.274306 |
0.274306 |
1
|
79
|
|
GO:0048609
|
multicellular organismal reproductive process
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0032504
|
multicellular organism reproduction
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0016071
|
mRNA metabolic process
|
0.284635 |
0.284635 |
1
|
339
|
|
GO:0009953
|
dorsal/ventral pattern formation
|
0.286561 |
0.286561 |
1
|
145
|
|
GO:0015631
|
tubulin binding
|
0.309091 |
0.309091 |
1
|
68
|
|
GO:0048869
|
cellular developmental process
|
0.322329 |
0.322329 |
1
|
1276
|
|
GO:0003723
|
RNA binding
|
0.351482 |
0.351482 |
1
|
652
|
|
GO:0000166
|
nucleotide binding
|
0.374077 |
0.374077 |
1
|
1178
|
|
GO:0009987
|
cellular process
|
0.381822 |
0.381822 |
2
|
6560
|
|
GO:0009108
|
coenzyme biosynthetic process
|
0.383178 |
0.383178 |
1
|
41
|
|
GO:0009058
|
biosynthetic process
|
0.402387 |
0.402387 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.402965 |
0.402965 |
1
|
2093
|
|
GO:0044430
|
cytoskeletal part
|
0.409756 |
0.409756 |
1
|
512
|
|
GO:0016070
|
RNA metabolic process
|
0.413829 |
0.413829 |
1
|
1191
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.415756 |
0.415756 |
1
|
1789
|
|
GO:0009798
|
axis specification
|
0.432030 |
0.432030 |
1
|
232
|
|
GO:0032502
|
developmental process
|
0.435255 |
0.435255 |
1
|
2638
|
|
GO:0005856
|
cytoskeleton
|
0.457213 |
0.457213 |
1
|
561
|
|
GO:0048468
|
cell development
|
0.457419 |
0.457419 |
1
|
1042
|
|
GO:0044428
|
nuclear part
|
0.469235 |
0.469235 |
1
|
830
|
|
GO:0048599
|
oocyte development
|
0.473016 |
0.473016 |
1
|
149
|
|
GO:0005575
|
cellular_component
|
0.478421 |
0.478421 |
2
|
8817
|
|
GO:0005623
|
cell
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.498346 |
0.498346 |
1
|
1959
|
|
GO:0044424
|
intracellular part
|
0.500760 |
0.500760 |
2
|
4384
|
|
GO:0005515
|
protein binding
|
0.518366 |
0.518366 |
1
|
1726
|
|
GO:0044249
|
cellular biosynthetic process
|
0.526807 |
0.526807 |
1
|
2034
|
|
GO:0032501
|
multicellular organismal process
|
0.527040 |
0.527040 |
1
|
3315
|
|
GO:0010467
|
gene expression
|
0.531494 |
0.531494 |
1
|
1907
|
|
GO:0003676
|
nucleic acid binding
|
0.549587 |
0.549587 |
1
|
1855
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.573530 |
0.573530 |
1
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.574428 |
0.574428 |
1
|
1227
|
|
GO:0005622
|
intracellular
|
0.583919 |
0.583919 |
2
|
4734
|
|
GO:0003824
|
catalytic activity
|
0.604523 |
0.604523 |
1
|
4106
|
|
GO:0016879
|
ligase activity, forming carbon-nitrogen bonds
|
0.610390 |
0.610390 |
1
|
141
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.639129 |
0.639129 |
1
|
2878
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0043229
|
intracellular organelle
|
0.646541 |
0.646541 |
2
|
3530
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0008380
|
RNA splicing
|
0.669082 |
0.669082 |
1
|
277
|
|
GO:0044444
|
cytoplasmic part
|
0.669378 |
0.669378 |
1
|
1863
|
|
GO:0071013
|
catalytic step 2 spliceosome
|
0.683333 |
0.683333 |
1
|
123
|
|
GO:0043170
|
macromolecule metabolic process
|
0.690797 |
0.690797 |
1
|
3588
|
|
GO:0008150
|
biological_process
|
0.693584 |
0.693584 |
2
|
10616
|
|
GO:0007292
|
female gamete generation
|
0.727380 |
0.727380 |
1
|
619
|
|
GO:0006397
|
mRNA processing
|
0.734411 |
0.734411 |
1
|
318
|
|
GO:0008152
|
metabolic process
|
0.739170 |
0.739170 |
1
|
5194
|
|
GO:0044237
|
cellular metabolic process
|
0.740940 |
0.740940 |
1
|
3814
|
|
GO:0032553
|
ribonucleotide binding
|
0.746180 |
0.746180 |
1
|
879
|
|
GO:0030272
|
5-formyltetrahydrofolate cyclo-ligase activity
|
0.750000 |
0.750000 |
1
|
3
|
|
GO:0003674
|
molecular_function
|
0.753361 |
0.753361 |
2
|
11064
|
|
GO:0032559
|
adenyl ribonucleotide binding
|
0.757705 |
0.757705 |
1
|
713
|
|
GO:0005737
|
cytoplasm
|
0.790684 |
0.790684 |
1
|
2378
|
|
GO:0015630
|
microtubule cytoskeleton
|
0.798574 |
0.798574 |
1
|
448
|
|
GO:0017076
|
purine nucleotide binding
|
0.803056 |
0.803056 |
1
|
946
|
|
GO:0030554
|
adenyl nucleotide binding
|
0.819239 |
0.819239 |
1
|
775
|
|
GO:0044238
|
primary metabolic process
|
0.819985 |
0.819985 |
1
|
4259
|
|
GO:0071011
|
precatalytic spliceosome
|
0.822222 |
0.822222 |
1
|
148
|
|
GO:0000398
|
nuclear mRNA splicing, via spliceosome
|
0.836478 |
0.836478 |
1
|
266
|
|
GO:0006732
|
coenzyme metabolic process
|
0.851852 |
0.851852 |
1
|
92
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0019953
|
sexual reproduction
|
0.859100 |
0.859100 |
1
|
878
|
|
GO:0007276
|
gamete generation
|
0.883697 |
0.883697 |
1
|
851
|
|
GO:0043234
|
protein complex
|
0.925620 |
0.925620 |
1
|
1564
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.929175 |
0.929175 |
1
|
879
|
|
GO:0008017
|
microtubule binding
|
0.941176 |
0.941176 |
1
|
64
|
|
GO:0003002
|
regionalization
|
0.942272 |
0.942272 |
1
|
506
|
|
GO:0043227
|
membrane-bounded organelle
|
0.963300 |
0.963300 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.963413 |
0.963413 |
1
|
2856
|
|
GO:0000375
|
RNA splicing, via transesterification reactions
|
0.963899 |
0.963899 |
1
|
267
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0048477
|
oogenesis
|
0.987076 |
0.987076 |
1
|
611
|
|
GO:0001883
|
purine nucleoside binding
|
0.992347 |
0.992347 |
1
|
778
|
|
GO:0000377
|
RNA splicing, via transesterification reactions with bulged adenosine as nucleophile
|
0.996255 |
0.996255 |
1
|
266
|
|
GO:0005524
|
ATP binding
|
0.998597 |
0.998597 |
1
|
712
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
2
|
12747
|