| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
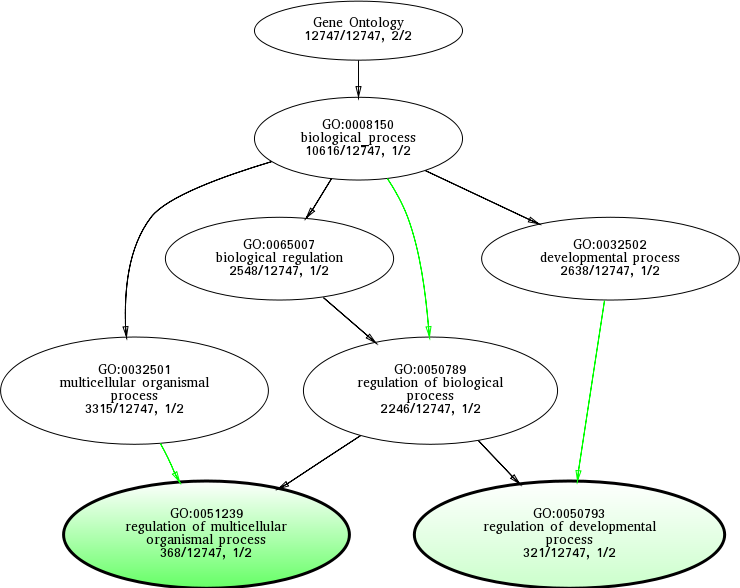
|
GO:0051239
|
regulation of multicellular organismal process
|
0.0844424 |
0.0844424 |
1
|
368
|
|
GO:0050793
|
regulation of developmental process
|
0.0844959 |
0.0844959 |
1
|
321
|
|
GO:0005783
|
endoplasmic reticulum
|
0.108098 |
0.108098 |
1
|
199
|
|
GO:2000026
|
regulation of multicellular organismal development
|
0.109792 |
0.109792 |
1
|
259
|
|
GO:0061061
|
muscle structure development
|
0.111258 |
0.111258 |
1
|
252
|
|
GO:0048634
|
regulation of muscle organ development
|
0.116580 |
0.116580 |
1
|
45
|
|
GO:0007517
|
muscle organ development
|
0.149102 |
0.149102 |
1
|
191
|
|
GO:0043226
|
organelle
|
0.160900 |
0.160900 |
2
|
3537
|
|
GO:0060537
|
muscle tissue development
|
0.163375 |
0.163375 |
1
|
91
|
|
GO:0050789
|
regulation of biological process
|
0.211567 |
0.211567 |
1
|
2246
|
|
GO:0065007
|
biological regulation
|
0.240015 |
0.240015 |
1
|
2548
|
|
GO:0009888
|
tissue development
|
0.245916 |
0.245916 |
1
|
557
|
|
GO:0032502
|
developmental process
|
0.248493 |
0.248493 |
1
|
2638
|
|
GO:0001882
|
nucleoside binding
|
0.258670 |
0.258670 |
1
|
784
|
|
GO:0005488
|
binding
|
0.259926 |
0.259926 |
2
|
5641
|
|
GO:0005737
|
cytoplasm
|
0.294170 |
0.294170 |
2
|
2378
|
|
GO:0016817
|
hydrolase activity, acting on acid anhydrides
|
0.301217 |
0.301217 |
1
|
594
|
|
GO:0032501
|
multicellular organismal process
|
0.312265 |
0.312265 |
1
|
3315
|
|
GO:0000166
|
nucleotide binding
|
0.374077 |
0.374077 |
1
|
1178
|
|
GO:0016202
|
regulation of striated muscle tissue development
|
0.395833 |
0.395833 |
1
|
38
|
|
GO:0043167
|
ion binding
|
0.460626 |
0.460626 |
1
|
1498
|
|
GO:0005575
|
cellular_component
|
0.478421 |
0.478421 |
2
|
8817
|
|
GO:0016787
|
hydrolase activity
|
0.480273 |
0.480273 |
1
|
1972
|
|
GO:0005623
|
cell
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044424
|
intracellular part
|
0.500760 |
0.500760 |
2
|
4384
|
|
GO:0005515
|
protein binding
|
0.518366 |
0.518366 |
1
|
1726
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0005622
|
intracellular
|
0.583919 |
0.583919 |
2
|
4734
|
|
GO:0003824
|
catalytic activity
|
0.604523 |
0.604523 |
1
|
4106
|
|
GO:0016887
|
ATPase activity
|
0.609672 |
0.609672 |
1
|
353
|
|
GO:0043229
|
intracellular organelle
|
0.646541 |
0.646541 |
2
|
3530
|
|
GO:0043227
|
membrane-bounded organelle
|
0.653325 |
0.653325 |
2
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.653801 |
0.653801 |
2
|
2856
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0044444
|
cytoplasmic part
|
0.669378 |
0.669378 |
1
|
1863
|
|
GO:0032553
|
ribonucleotide binding
|
0.746180 |
0.746180 |
1
|
879
|
|
GO:0003674
|
molecular_function
|
0.753361 |
0.753361 |
2
|
11064
|
|
GO:0032559
|
adenyl ribonucleotide binding
|
0.757705 |
0.757705 |
1
|
713
|
|
GO:0008270
|
zinc ion binding
|
0.781440 |
0.781440 |
1
|
901
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0017076
|
purine nucleotide binding
|
0.803056 |
0.803056 |
1
|
946
|
|
GO:0030554
|
adenyl nucleotide binding
|
0.819239 |
0.819239 |
1
|
775
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0005634
|
nucleus
|
0.870016 |
0.870016 |
1
|
1826
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.929175 |
0.929175 |
1
|
879
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0008150
|
biological_process
|
0.972063 |
0.972063 |
1
|
10616
|
|
GO:0017111
|
nucleoside-triphosphatase activity
|
0.983022 |
0.983022 |
1
|
579
|
|
GO:0014706
|
striated muscle tissue development
|
0.989011 |
0.989011 |
1
|
90
|
|
GO:0016462
|
pyrophosphatase activity
|
0.991582 |
0.991582 |
1
|
589
|
|
GO:0001883
|
purine nucleoside binding
|
0.992347 |
0.992347 |
1
|
778
|
|
GO:0005524
|
ATP binding
|
0.998597 |
0.998597 |
1
|
712
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
2
|
12747
|
|
GO:0016818
|
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
|
1.00000 |
1.00000 |
1
|
594
|