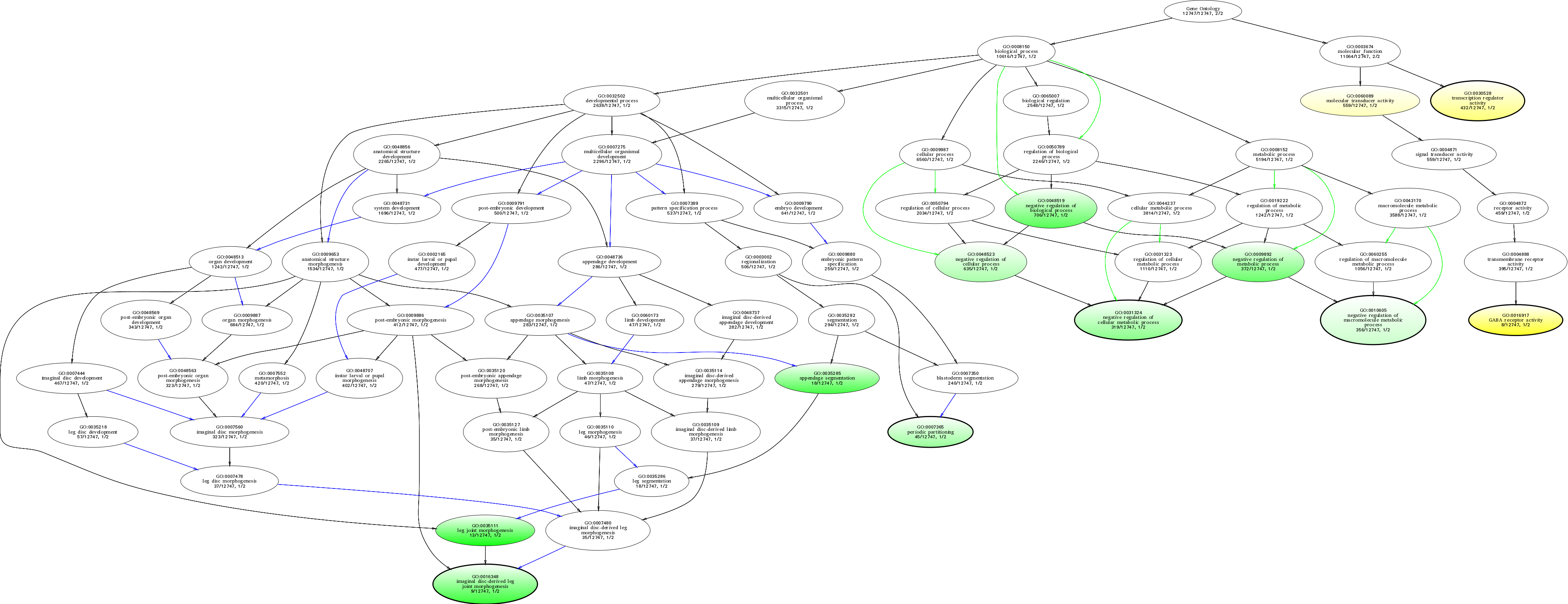
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0035111
|
leg joint morphogenesis
|
0.00847458 |
0.00847458 |
1
|
13
|
|
GO:0016917
|
GABA receptor activity
|
0.0202532 |
0.0202532 |
1
|
8
|
|
GO:0016348
|
imaginal disc-derived leg joint morphogenesis
|
0.0218447 |
0.0218447 |
1
|
9
|
|
GO:0035285
|
appendage segmentation
|
0.0347490 |
0.0347490 |
1
|
18
|
|
GO:0048519
|
negative regulation of biological process
|
0.0665034 |
0.0665034 |
1
|
706
|
|
GO:0009892
|
negative regulation of metabolic process
|
0.0691964 |
0.0691964 |
1
|
372
|
|
GO:0030528
|
transcription regulator activity
|
0.0765699 |
0.0765699 |
1
|
432
|
|
GO:0031324
|
negative regulation of cellular metabolic process
|
0.0793138 |
0.0793138 |
1
|
319
|
|
GO:0007365
|
periodic partitioning
|
0.0889328 |
0.0889328 |
1
|
45
|
|
GO:0048523
|
negative regulation of cellular process
|
0.0966514 |
0.0966514 |
1
|
635
|
|
GO:0060089
|
molecular transducer activity
|
0.0985001 |
0.0985001 |
1
|
559
|
|
GO:0010605
|
negative regulation of macromolecule metabolic process
|
0.0988340 |
0.0988340 |
1
|
356
|
|
GO:0004965
|
GABA-B receptor activity
|
0.104167 |
0.104167 |
1
|
5
|
|
GO:0048736
|
appendage development
|
0.110896 |
0.110896 |
1
|
286
|
|
GO:0007478
|
leg disc morphogenesis
|
0.111782 |
0.111782 |
1
|
37
|
|
GO:0008066
|
glutamate receptor activity
|
0.113924 |
0.113924 |
1
|
45
|
|
GO:0051172
|
negative regulation of nitrogen compound metabolic process
|
0.114943 |
0.114943 |
1
|
250
|
|
GO:0035218
|
leg disc development
|
0.122056 |
0.122056 |
1
|
57
|
|
GO:0035127
|
post-embryonic limb morphogenesis
|
0.128205 |
0.128205 |
1
|
35
|
|
GO:0009890
|
negative regulation of biosynthetic process
|
0.129715 |
0.129715 |
1
|
282
|
|
GO:0035109
|
imaginal disc-derived limb morphogenesis
|
0.132143 |
0.132143 |
1
|
37
|
|
GO:0045934
|
negative regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.135355 |
0.135355 |
1
|
250
|
|
GO:0031327
|
negative regulation of cellular biosynthetic process
|
0.136166 |
0.136166 |
1
|
282
|
|
GO:0010629
|
negative regulation of gene expression
|
0.149146 |
0.149146 |
1
|
288
|
|
GO:0051253
|
negative regulation of RNA metabolic process
|
0.155744 |
0.155744 |
1
|
202
|
|
GO:0060173
|
limb development
|
0.164336 |
0.164336 |
1
|
47
|
|
GO:0010558
|
negative regulation of macromolecule biosynthetic process
|
0.165094 |
0.165094 |
1
|
280
|
|
GO:0035108
|
limb morphogenesis
|
0.166078 |
0.166078 |
1
|
47
|
|
GO:2000113
|
negative regulation of cellular macromolecule biosynthetic process
|
0.172946 |
0.172946 |
1
|
280
|
|
GO:0007366
|
periodic partitioning by pair rule gene
|
0.177778 |
0.177778 |
1
|
8
|
|
GO:0035107
|
appendage morphogenesis
|
0.184365 |
0.184365 |
1
|
283
|
|
GO:0009791
|
post-embryonic development
|
0.189538 |
0.189538 |
1
|
500
|
|
GO:0007389
|
pattern specification process
|
0.203563 |
0.203563 |
1
|
537
|
|
GO:0019222
|
regulation of metabolic process
|
0.210401 |
0.210401 |
1
|
1242
|
|
GO:0050789
|
regulation of biological process
|
0.211567 |
0.211567 |
1
|
2246
|
|
GO:0016564
|
transcription repressor activity
|
0.215278 |
0.215278 |
1
|
93
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.237215 |
0.237215 |
1
|
1039
|
|
GO:0065007
|
biological regulation
|
0.240015 |
0.240015 |
1
|
2548
|
|
GO:0009790
|
embryo development
|
0.242987 |
0.242987 |
1
|
641
|
|
GO:0016481
|
negative regulation of transcription
|
0.243724 |
0.243724 |
1
|
233
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.244709 |
0.244709 |
1
|
1110
|
|
GO:0032502
|
developmental process
|
0.248493 |
0.248493 |
1
|
2638
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.259446 |
0.259446 |
1
|
412
|
|
GO:0043167
|
ion binding
|
0.265556 |
0.265556 |
1
|
1498
|
|
GO:0045892
|
negative regulation of transcription, DNA-dependent
|
0.266030 |
0.266030 |
1
|
195
|
|
GO:0007552
|
metamorphosis
|
0.273794 |
0.273794 |
1
|
420
|
|
GO:0048569
|
post-embryonic organ development
|
0.276167 |
0.276167 |
1
|
343
|
|
GO:0006357
|
regulation of transcription from RNA polymerase II promoter
|
0.282934 |
0.282934 |
1
|
189
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.283034 |
0.283034 |
1
|
1056
|
|
GO:0032774
|
RNA biosynthetic process
|
0.290017 |
0.290017 |
1
|
703
|
|
GO:0003704
|
specific RNA polymerase II transcription factor activity
|
0.292135 |
0.292135 |
1
|
78
|
|
GO:0009880
|
embryonic pattern specification
|
0.294591 |
0.294591 |
1
|
256
|
|
GO:0050794
|
regulation of cellular process
|
0.306371 |
0.306371 |
1
|
2034
|
|
GO:0032501
|
multicellular organismal process
|
0.312265 |
0.312265 |
1
|
3315
|
|
GO:0003676
|
nucleic acid binding
|
0.328842 |
0.328842 |
1
|
1855
|
|
GO:0006366
|
transcription from RNA polymerase II promoter
|
0.360913 |
0.360913 |
1
|
253
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.366791 |
0.366791 |
1
|
888
|
|
GO:0009887
|
organ morphogenesis
|
0.374384 |
0.374384 |
1
|
684
|
|
GO:0007444
|
imaginal disc development
|
0.376006 |
0.376006 |
1
|
467
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.381695 |
0.381695 |
1
|
905
|
|
GO:0006350
|
transcription
|
0.387810 |
0.387810 |
1
|
859
|
|
GO:0035286
|
leg segmentation
|
0.391304 |
0.391304 |
1
|
18
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.396252 |
0.396252 |
1
|
888
|
|
GO:0043226
|
organelle
|
0.401157 |
0.401157 |
1
|
3537
|
|
GO:0009058
|
biosynthetic process
|
0.402387 |
0.402387 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.402965 |
0.402965 |
1
|
2093
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.405649 |
0.405649 |
1
|
1623
|
|
GO:0016070
|
RNA metabolic process
|
0.413829 |
0.413829 |
1
|
1191
|
|
GO:0048563
|
post-embryonic organ morphogenesis
|
0.415167 |
0.415167 |
1
|
323
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.415756 |
0.415756 |
1
|
1789
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.440308 |
0.440308 |
1
|
686
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.451726 |
0.451726 |
1
|
903
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.456989 |
0.456989 |
1
|
850
|
|
GO:0008152
|
metabolic process
|
0.489261 |
0.489261 |
1
|
5194
|
|
GO:0010468
|
regulation of gene expression
|
0.489930 |
0.489930 |
1
|
973
|
|
GO:0044237
|
cellular metabolic process
|
0.490989 |
0.490989 |
1
|
3814
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.496927 |
0.496927 |
1
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.498346 |
0.498346 |
1
|
1959
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.513595 |
0.513595 |
1
|
850
|
|
GO:0044249
|
cellular biosynthetic process
|
0.526807 |
0.526807 |
1
|
2034
|
|
GO:0010467
|
gene expression
|
0.531494 |
0.531494 |
1
|
1907
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0007560
|
imaginal disc morphogenesis
|
0.575758 |
0.575758 |
1
|
323
|
|
GO:0035282
|
segmentation
|
0.581028 |
0.581028 |
1
|
294
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0009987
|
cellular process
|
0.617935 |
0.617935 |
1
|
6560
|
|
GO:0003702
|
RNA polymerase II transcription factor activity
|
0.618056 |
0.618056 |
1
|
267
|
|
GO:0035120
|
post-embryonic appendage morphogenesis
|
0.638095 |
0.638095 |
1
|
268
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.639129 |
0.639129 |
1
|
2878
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.684327 |
0.684327 |
1
|
620
|
|
GO:0045449
|
regulation of transcription
|
0.687166 |
0.687166 |
1
|
771
|
|
GO:0043170
|
macromolecule metabolic process
|
0.690797 |
0.690797 |
1
|
3588
|
|
GO:0005623
|
cell
|
0.702620 |
0.702620 |
1
|
6195
|
|
GO:0044464
|
cell part
|
0.702620 |
0.702620 |
1
|
6195
|
|
GO:0044424
|
intracellular part
|
0.707667 |
0.707667 |
1
|
4384
|
|
GO:0007480
|
imaginal disc-derived leg morphogenesis
|
0.729167 |
0.729167 |
1
|
35
|
|
GO:0003674
|
molecular_function
|
0.753361 |
0.753361 |
2
|
11064
|
|
GO:0005488
|
binding
|
0.759777 |
0.759777 |
1
|
5641
|
|
GO:0005622
|
intracellular
|
0.764165 |
0.764165 |
1
|
4734
|
|
GO:0007350
|
blastoderm segmentation
|
0.779221 |
0.779221 |
1
|
240
|
|
GO:0008270
|
zinc ion binding
|
0.781440 |
0.781440 |
1
|
901
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0043229
|
intracellular organelle
|
0.804100 |
0.804100 |
1
|
3530
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.814170 |
0.814170 |
1
|
701
|
|
GO:0044238
|
primary metabolic process
|
0.819985 |
0.819985 |
1
|
4259
|
|
GO:0004872
|
receptor activity
|
0.821109 |
0.821109 |
1
|
459
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.842767 |
0.842767 |
1
|
402
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0004888
|
transmembrane receptor activity
|
0.860566 |
0.860566 |
1
|
395
|
|
GO:0005575
|
cellular_component
|
0.904963 |
0.904963 |
1
|
8817
|
|
GO:0003002
|
regionalization
|
0.942272 |
0.942272 |
1
|
506
|
|
GO:0002165
|
instar larval or pupal development
|
0.954000 |
0.954000 |
1
|
477
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0008150
|
biological_process
|
0.972063 |
0.972063 |
1
|
10616
|
|
GO:0035114
|
imaginal disc-derived appendage morphogenesis
|
0.975524 |
0.975524 |
1
|
279
|
|
GO:0035110
|
leg morphogenesis
|
0.978723 |
0.978723 |
1
|
46
|
|
GO:0048737
|
imaginal disc-derived appendage development
|
0.986014 |
0.986014 |
1
|
282
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
2
|
12747
|
|
GO:0004871
|
signal transducer activity
|
1.00000 |
1.00000 |
1
|
559
|