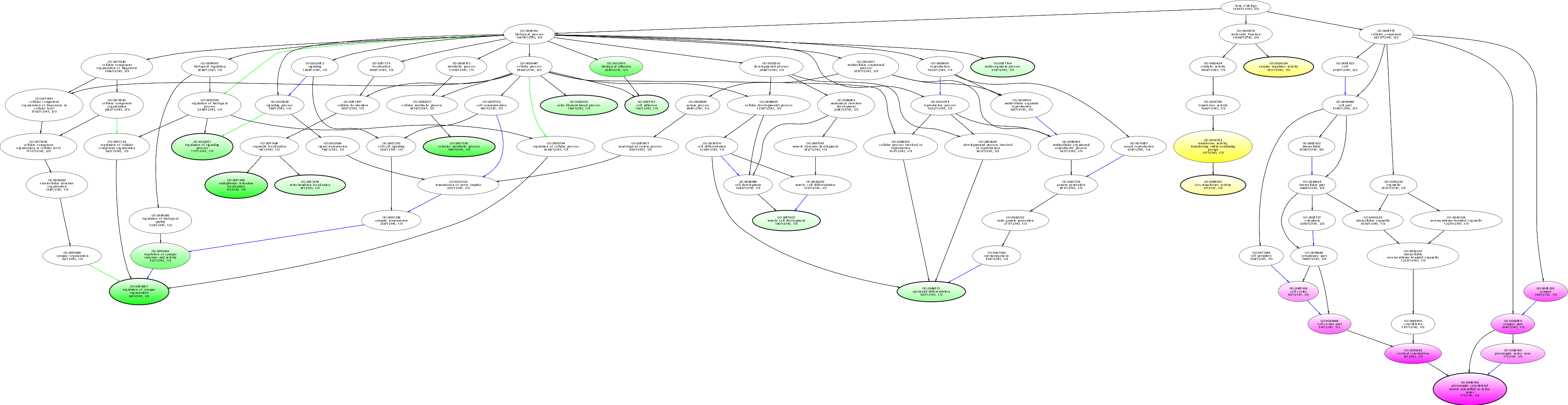
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0048788
|
presynaptic cytoskeletal matrix assembled at active zones
|
0.00900901 |
0.00900901 |
1
|
1
|
|
GO:0030863
|
cortical cytoskeleton
|
0.0153061 |
0.0153061 |
1
|
9
|
|
GO:0050807
|
regulation of synapse organization
|
0.0184466 |
0.0184466 |
1
|
38
|
|
GO:0051643
|
endoplasmic reticulum localization
|
0.0215054 |
0.0215054 |
1
|
2
|
|
GO:0016782
|
transferase activity, transferring sulfur-containing groups
|
0.0262136 |
0.0262136 |
1
|
27
|
|
GO:0051186
|
cofactor metabolic process
|
0.0283167 |
0.0283167 |
1
|
108
|
|
GO:0044456
|
synapse part
|
0.0349744 |
0.0349744 |
1
|
104
|
|
GO:0045202
|
synapse
|
0.0449093 |
0.0449093 |
1
|
134
|
|
GO:0022610
|
biological adhesion
|
0.0570970 |
0.0570970 |
1
|
206
|
|
GO:0044448
|
cell cortex part
|
0.0613121 |
0.0613121 |
1
|
58
|
|
GO:0030234
|
enzyme regulator activity
|
0.0624454 |
0.0624454 |
1
|
351
|
|
GO:0050803
|
regulation of synapse structure and activity
|
0.0668380 |
0.0668380 |
1
|
52
|
|
GO:0048786
|
presynaptic active zone
|
0.0673077 |
0.0673077 |
1
|
7
|
|
GO:0023051
|
regulation of signaling process
|
0.0723656 |
0.0723656 |
1
|
171
|
|
GO:0005938
|
cell cortex
|
0.0724804 |
0.0724804 |
1
|
89
|
|
GO:0008410
|
CoA-transferase activity
|
0.0740741 |
0.0740741 |
1
|
2
|
|
GO:0048515
|
spermatid differentiation
|
0.0741002 |
0.0741002 |
1
|
105
|
|
GO:0030029
|
actin filament-based process
|
0.0826842 |
0.0826842 |
1
|
186
|
|
GO:0007155
|
cell adhesion
|
0.0851090 |
0.0851090 |
1
|
192
|
|
GO:0051646
|
mitochondrion localization
|
0.0860215 |
0.0860215 |
1
|
8
|
|
GO:0051704
|
multi-organism process
|
0.0874732 |
0.0874732 |
1
|
319
|
|
GO:0055001
|
muscle cell development
|
0.0938967 |
0.0938967 |
1
|
100
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.102602 |
0.102602 |
2
|
2108
|
|
GO:0042692
|
muscle cell differentiation
|
0.102644 |
0.102644 |
1
|
132
|
|
GO:0061061
|
muscle structure development
|
0.111258 |
0.111258 |
1
|
252
|
|
GO:0030239
|
myofibril assembly
|
0.117886 |
0.117886 |
1
|
29
|
|
GO:0035466
|
regulation of signaling pathway
|
0.126661 |
0.126661 |
1
|
324
|
|
GO:0010927
|
cellular component assembly involved in morphogenesis
|
0.134157 |
0.134157 |
1
|
152
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.142259 |
0.142259 |
2
|
1562
|
|
GO:0051640
|
organelle localization
|
0.153213 |
0.153213 |
1
|
93
|
|
GO:0051128
|
regulation of cellular component organization
|
0.156775 |
0.156775 |
1
|
282
|
|
GO:0043062
|
extracellular structure organization
|
0.168915 |
0.168915 |
1
|
134
|
|
GO:0048646
|
anatomical structure formation involved in morphogenesis
|
0.176649 |
0.176649 |
1
|
466
|
|
GO:0044444
|
cytoplasmic part
|
0.180530 |
0.180530 |
2
|
1863
|
|
GO:0007267
|
cell-cell signaling
|
0.188801 |
0.188801 |
1
|
263
|
|
GO:0019226
|
transmission of nerve impulse
|
0.194826 |
0.194826 |
1
|
241
|
|
GO:0007154
|
cell communication
|
0.196383 |
0.196383 |
1
|
461
|
|
GO:0023046
|
signaling process
|
0.199779 |
0.199779 |
1
|
760
|
|
GO:0003006
|
developmental process involved in reproduction
|
0.204901 |
0.204901 |
1
|
602
|
|
GO:0051705
|
behavioral interaction between organisms
|
0.209431 |
0.209431 |
1
|
151
|
|
GO:0007281
|
germ cell development
|
0.219828 |
0.219828 |
1
|
306
|
|
GO:0051056
|
regulation of small GTPase mediated signal transduction
|
0.219858 |
0.219858 |
1
|
93
|
|
GO:0032501
|
multicellular organismal process
|
0.231607 |
0.231607 |
2
|
3315
|
|
GO:0009987
|
cellular process
|
0.235914 |
0.235914 |
3
|
6560
|
|
GO:0051641
|
cellular localization
|
0.242560 |
0.242560 |
1
|
607
|
|
GO:0048610
|
cellular process involved in reproduction
|
0.248418 |
0.248418 |
1
|
613
|
|
GO:0005811
|
lipid particle
|
0.250439 |
0.250439 |
1
|
250
|
|
GO:0016740
|
transferase activity
|
0.250852 |
0.250852 |
1
|
1030
|
|
GO:0005088
|
Ras guanyl-nucleotide exchange factor activity
|
0.251969 |
0.251969 |
1
|
32
|
|
GO:0007165
|
signal transduction
|
0.254057 |
0.254057 |
1
|
548
|
|
GO:0048232
|
male gamete generation
|
0.254994 |
0.254994 |
1
|
217
|
|
GO:0000003
|
reproduction
|
0.261920 |
0.261920 |
1
|
1022
|
|
GO:0022414
|
reproductive process
|
0.261920 |
0.261920 |
1
|
1022
|
|
GO:0007266
|
Rho protein signal transduction
|
0.280000 |
0.280000 |
1
|
28
|
|
GO:0065008
|
regulation of biological quality
|
0.286107 |
0.286107 |
1
|
729
|
|
GO:0005737
|
cytoplasm
|
0.294170 |
0.294170 |
2
|
2378
|
|
GO:0031032
|
actomyosin structure organization
|
0.306011 |
0.306011 |
1
|
56
|
|
GO:0071944
|
cell periphery
|
0.311271 |
0.311271 |
1
|
724
|
|
GO:0009966
|
regulation of signal transduction
|
0.312044 |
0.312044 |
1
|
171
|
|
GO:0035023
|
regulation of Rho protein signal transduction
|
0.320988 |
0.320988 |
1
|
26
|
|
GO:0007286
|
spermatid development
|
0.330097 |
0.330097 |
1
|
102
|
|
GO:0005575
|
cellular_component
|
0.330897 |
0.330897 |
3
|
8817
|
|
GO:0023052
|
signaling
|
0.338079 |
0.338079 |
1
|
1364
|
|
GO:0030036
|
actin cytoskeleton organization
|
0.339518 |
0.339518 |
1
|
183
|
|
GO:0005623
|
cell
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0044464
|
cell part
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.346904 |
0.346904 |
1
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.347592 |
0.347592 |
1
|
1227
|
|
GO:0032989
|
cellular component morphogenesis
|
0.348979 |
0.348979 |
1
|
566
|
|
GO:0045214
|
sarcomere organization
|
0.357143 |
0.357143 |
1
|
20
|
|
GO:0003008
|
system process
|
0.360531 |
0.360531 |
1
|
664
|
|
GO:0007610
|
behavior
|
0.361111 |
0.361111 |
1
|
559
|
|
GO:0051647
|
nucleus localization
|
0.376344 |
0.376344 |
1
|
35
|
|
GO:0050896
|
response to stimulus
|
0.376795 |
0.376795 |
1
|
1548
|
|
GO:0016337
|
cell-cell adhesion
|
0.385417 |
0.385417 |
1
|
74
|
|
GO:0002118
|
aggressive behavior
|
0.397351 |
0.397351 |
1
|
60
|
|
GO:0060589
|
nucleoside-triphosphatase regulator activity
|
0.410256 |
0.410256 |
1
|
144
|
|
GO:0005085
|
guanyl-nucleotide exchange factor activity
|
0.415493 |
0.415493 |
1
|
59
|
|
GO:0023034
|
intracellular signaling pathway
|
0.430962 |
0.430962 |
1
|
412
|
|
GO:0048869
|
cellular developmental process
|
0.442161 |
0.442161 |
1
|
1276
|
|
GO:0006084
|
acetyl-CoA metabolic process
|
0.445652 |
0.445652 |
1
|
41
|
|
GO:0051179
|
localization
|
0.445834 |
0.445834 |
1
|
1896
|
|
GO:0005622
|
intracellular
|
0.446165 |
0.446165 |
3
|
4734
|
|
GO:0007010
|
cytoskeleton organization
|
0.453469 |
0.453469 |
1
|
536
|
|
GO:0005856
|
cytoskeleton
|
0.457213 |
0.457213 |
1
|
561
|
|
GO:0048468
|
cell development
|
0.457419 |
0.457419 |
1
|
1042
|
|
GO:0022607
|
cellular component assembly
|
0.475076 |
0.475076 |
1
|
577
|
|
GO:0048609
|
multicellular organismal reproductive process
|
0.480389 |
0.480389 |
1
|
937
|
|
GO:0032504
|
multicellular organism reproduction
|
0.480389 |
0.480389 |
1
|
937
|
|
GO:0035556
|
intracellular signal transduction
|
0.499208 |
0.499208 |
1
|
315
|
|
GO:0008411
|
4-hydroxybutyrate CoA-transferase activity
|
0.500000 |
0.500000 |
1
|
1
|
|
GO:0044085
|
cellular component biogenesis
|
0.509834 |
0.509834 |
1
|
632
|
|
GO:0050789
|
regulation of biological process
|
0.509927 |
0.509927 |
1
|
2246
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.524233 |
0.524233 |
2
|
1517
|
|
GO:0007265
|
Ras protein signal transduction
|
0.540541 |
0.540541 |
1
|
100
|
|
GO:0065007
|
biological regulation
|
0.561089 |
0.561089 |
1
|
2548
|
|
GO:0032502
|
developmental process
|
0.575616 |
0.575616 |
1
|
2638
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0007264
|
small GTPase mediated signal transduction
|
0.587302 |
0.587302 |
1
|
185
|
|
GO:0003824
|
catalytic activity
|
0.604523 |
0.604523 |
1
|
4106
|
|
GO:0071844
|
cellular component assembly at cellular level
|
0.607267 |
0.607267 |
1
|
568
|
|
GO:0050794
|
regulation of cellular process
|
0.666348 |
0.666348 |
1
|
2034
|
|
GO:0050808
|
synapse organization
|
0.686567 |
0.686567 |
1
|
92
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0046578
|
regulation of Ras protein signal transduction
|
0.705357 |
0.705357 |
1
|
79
|
|
GO:0005083
|
small GTPase regulator activity
|
0.746479 |
0.746479 |
1
|
106
|
|
GO:0005089
|
Rho guanyl-nucleotide exchange factor activity
|
0.750000 |
0.750000 |
1
|
24
|
|
GO:0043226
|
organelle
|
0.785296 |
0.785296 |
1
|
3537
|
|
GO:0044424
|
intracellular part
|
0.793631 |
0.793631 |
2
|
4384
|
|
GO:0055002
|
striated muscle cell development
|
0.798387 |
0.798387 |
1
|
99
|
|
GO:0006732
|
coenzyme metabolic process
|
0.851852 |
0.851852 |
1
|
92
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0019953
|
sexual reproduction
|
0.859100 |
0.859100 |
1
|
878
|
|
GO:0007268
|
synaptic transmission
|
0.866667 |
0.866667 |
1
|
234
|
|
GO:0008152
|
metabolic process
|
0.866808 |
0.866808 |
1
|
5194
|
|
GO:0044237
|
cellular metabolic process
|
0.868168 |
0.868168 |
1
|
3814
|
|
GO:0007276
|
gamete generation
|
0.883697 |
0.883697 |
1
|
851
|
|
GO:0051146
|
striated muscle cell differentiation
|
0.939394 |
0.939394 |
1
|
124
|
|
GO:0016043
|
cellular component organization
|
0.947563 |
0.947563 |
2
|
2052
|
|
GO:0006996
|
organelle organization
|
0.951347 |
0.951347 |
1
|
1182
|
|
GO:0003674
|
molecular_function
|
0.952326 |
0.952326 |
2
|
11064
|
|
GO:0050877
|
neurological system process
|
0.953313 |
0.953313 |
1
|
633
|
|
GO:0043229
|
intracellular organelle
|
0.961659 |
0.961659 |
1
|
3530
|
|
GO:0002121
|
inter-male aggressive behavior
|
0.966667 |
0.966667 |
1
|
58
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0030695
|
GTPase regulator activity
|
0.986111 |
0.986111 |
1
|
142
|
|
GO:0007283
|
spermatogenesis
|
0.995392 |
0.995392 |
1
|
216
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|