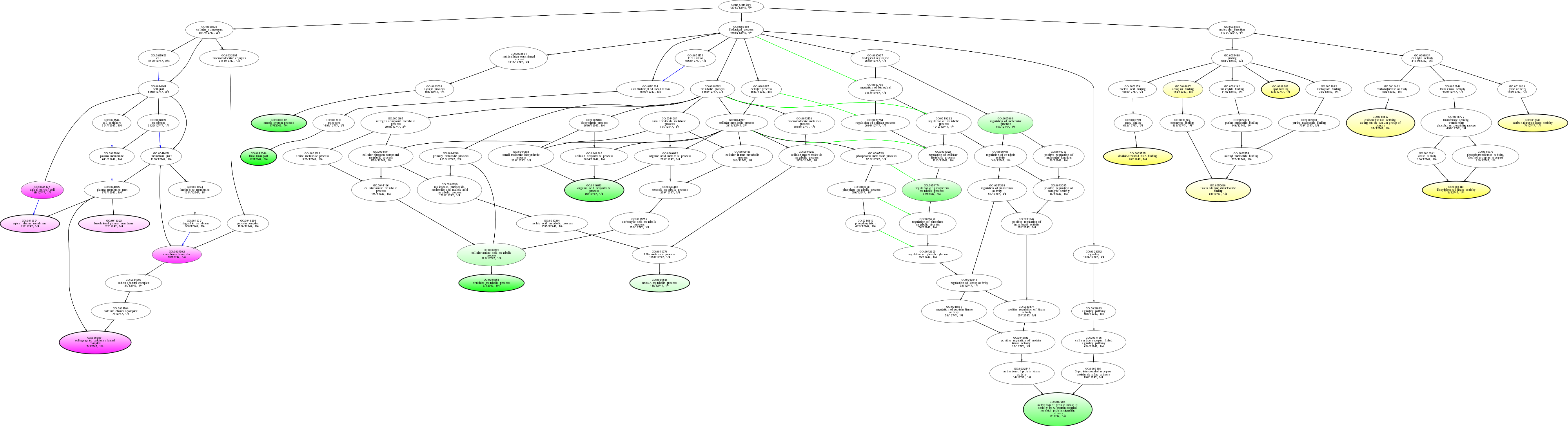
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0042044
|
fluid transport
|
0.00801603 |
0.00801603 |
1
|
12
|
|
GO:0006591
|
ornithine metabolic process
|
0.0174419 |
0.0174419 |
1
|
3
|
|
GO:0005891
|
voltage-gated calcium channel complex
|
0.0187668 |
0.0187668 |
1
|
7
|
|
GO:0045177
|
apical part of cell
|
0.0205567 |
0.0205567 |
1
|
64
|
|
GO:0034702
|
ion channel complex
|
0.0210151 |
0.0210151 |
1
|
53
|
|
GO:0004143
|
diacylglycerol kinase activity
|
0.0226131 |
0.0226131 |
1
|
9
|
|
GO:0003012
|
muscle system process
|
0.0256024 |
0.0256024 |
1
|
17
|
|
GO:0016053
|
organic acid biosynthetic process
|
0.0301205 |
0.0301205 |
1
|
65
|
|
GO:0003725
|
double-stranded RNA binding
|
0.0306748 |
0.0306748 |
1
|
20
|
|
GO:0007205
|
activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway
|
0.0441176 |
0.0441176 |
1
|
9
|
|
GO:0008289
|
lipid binding
|
0.0466029 |
0.0466029 |
1
|
133
|
|
GO:0051174
|
regulation of phosphorus metabolic process
|
0.0483153 |
0.0483153 |
1
|
76
|
|
GO:0016840
|
carbon-nitrogen lyase activity
|
0.0486111 |
0.0486111 |
1
|
7
|
|
GO:0016627
|
oxidoreductase activity, acting on the CH-CH group of donors
|
0.0579937 |
0.0579937 |
1
|
37
|
|
GO:0065009
|
regulation of molecular function
|
0.0655416 |
0.0655416 |
1
|
167
|
|
GO:0048037
|
cofactor binding
|
0.0665778 |
0.0665778 |
1
|
191
|
|
GO:0050660
|
flavin adenine dinucleotide binding
|
0.0715123 |
0.0715123 |
1
|
61
|
|
GO:0016324
|
apical plasma membrane
|
0.0719603 |
0.0719603 |
1
|
29
|
|
GO:0006520
|
cellular amino acid metabolic process
|
0.0784268 |
0.0784268 |
1
|
172
|
|
GO:0016323
|
basolateral plasma membrane
|
0.0991957 |
0.0991957 |
1
|
37
|
|
GO:0034660
|
ncRNA metabolic process
|
0.0999160 |
0.0999160 |
1
|
119
|
|
GO:0016829
|
lyase activity
|
0.101589 |
0.101589 |
1
|
144
|
|
GO:0009309
|
amine biosynthetic process
|
0.102119 |
0.102119 |
1
|
53
|
|
GO:0044283
|
small molecule biosynthetic process
|
0.105199 |
0.105199 |
1
|
259
|
|
GO:0017150
|
tRNA dihydrouridine synthase activity
|
0.108108 |
0.108108 |
1
|
4
|
|
GO:0050790
|
regulation of catalytic activity
|
0.119487 |
0.119487 |
1
|
149
|
|
GO:0006526
|
arginine biosynthetic process
|
0.125000 |
0.125000 |
1
|
2
|
|
GO:0019220
|
regulation of phosphate metabolic process
|
0.136691 |
0.136691 |
1
|
76
|
|
GO:0009987
|
cellular process
|
0.145754 |
0.145754 |
4
|
6560
|
|
GO:0003824
|
catalytic activity
|
0.147500 |
0.147500 |
3
|
4106
|
|
GO:0034470
|
ncRNA processing
|
0.148707 |
0.148707 |
1
|
69
|
|
GO:0072511
|
divalent inorganic cation transport
|
0.149798 |
0.149798 |
1
|
37
|
|
GO:0009064
|
glutamine family amino acid metabolic process
|
0.151163 |
0.151163 |
1
|
26
|
|
GO:0042325
|
regulation of phosphorylation
|
0.160839 |
0.160839 |
1
|
69
|
|
GO:0008652
|
cellular amino acid biosynthetic process
|
0.175355 |
0.175355 |
1
|
37
|
|
GO:0044271
|
cellular nitrogen compound biosynthetic process
|
0.177736 |
0.177736 |
1
|
252
|
|
GO:0044106
|
cellular amine metabolic process
|
0.178182 |
0.178182 |
1
|
195
|
|
GO:0005245
|
voltage-gated calcium channel activity
|
0.180328 |
0.180328 |
1
|
11
|
|
GO:0006082
|
organic acid metabolic process
|
0.189003 |
0.189003 |
1
|
259
|
|
GO:0006525
|
arginine metabolic process
|
0.192308 |
0.192308 |
1
|
5
|
|
GO:0019992
|
diacylglycerol binding
|
0.195489 |
0.195489 |
1
|
26
|
|
GO:0003008
|
system process
|
0.200302 |
0.200302 |
1
|
664
|
|
GO:0042180
|
cellular ketone metabolic process
|
0.203187 |
0.203187 |
1
|
280
|
|
GO:0006396
|
RNA processing
|
0.214286 |
0.214286 |
1
|
414
|
|
GO:0005262
|
calcium channel activity
|
0.218750 |
0.218750 |
1
|
28
|
|
GO:0071944
|
cell periphery
|
0.220095 |
0.220095 |
1
|
724
|
|
GO:0006811
|
ion transport
|
0.221109 |
0.221109 |
1
|
331
|
|
GO:0034704
|
calcium channel complex
|
0.225806 |
0.225806 |
1
|
7
|
|
GO:0070838
|
divalent metal ion transport
|
0.227848 |
0.227848 |
1
|
36
|
|
GO:0044459
|
plasma membrane part
|
0.236675 |
0.236675 |
1
|
373
|
|
GO:0009790
|
embryo development
|
0.242987 |
0.242987 |
1
|
641
|
|
GO:0055085
|
transmembrane transport
|
0.246988 |
0.246988 |
1
|
469
|
|
GO:0004056
|
argininosuccinate lyase activity
|
0.250000 |
0.250000 |
1
|
1
|
|
GO:0042450
|
arginine biosynthetic process via ornithine
|
0.250000 |
0.250000 |
1
|
1
|
|
GO:0046394
|
carboxylic acid biosynthetic process
|
0.250965 |
0.250965 |
1
|
65
|
|
GO:0007165
|
signal transduction
|
0.254057 |
0.254057 |
1
|
548
|
|
GO:0023046
|
signaling process
|
0.257083 |
0.257083 |
1
|
760
|
|
GO:0001882
|
nucleoside binding
|
0.258670 |
0.258670 |
1
|
784
|
|
GO:0009084
|
glutamine family amino acid biosynthetic process
|
0.265306 |
0.265306 |
1
|
13
|
|
GO:0055114
|
oxidation-reduction process
|
0.269455 |
0.269455 |
1
|
516
|
|
GO:0051347
|
positive regulation of transferase activity
|
0.271739 |
0.271739 |
1
|
25
|
|
GO:0005261
|
cation channel activity
|
0.272340 |
0.272340 |
1
|
128
|
|
GO:0005215
|
transporter activity
|
0.274270 |
0.274270 |
1
|
852
|
|
GO:0022803
|
passive transmembrane transporter activity
|
0.276860 |
0.276860 |
1
|
201
|
|
GO:0005244
|
voltage-gated ion channel activity
|
0.285714 |
0.285714 |
1
|
54
|
|
GO:0009308
|
amine metabolic process
|
0.286510 |
0.286510 |
1
|
325
|
|
GO:0005886
|
plasma membrane
|
0.292027 |
0.292027 |
1
|
641
|
|
GO:0008152
|
metabolic process
|
0.296531 |
0.296531 |
3
|
5194
|
|
GO:0008033
|
tRNA processing
|
0.296610 |
0.296610 |
1
|
35
|
|
GO:0044237
|
cellular metabolic process
|
0.299056 |
0.299056 |
3
|
3814
|
|
GO:0022838
|
substrate-specific channel activity
|
0.307448 |
0.307448 |
1
|
194
|
|
GO:0007186
|
G-protein coupled receptor protein signaling pathway
|
0.318910 |
0.318910 |
1
|
199
|
|
GO:0022843
|
voltage-gated cation channel activity
|
0.321168 |
0.321168 |
1
|
44
|
|
GO:0003723
|
RNA binding
|
0.351482 |
0.351482 |
1
|
652
|
|
GO:0051338
|
regulation of transferase activity
|
0.355705 |
0.355705 |
1
|
53
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.356248 |
0.356248 |
2
|
2093
|
|
GO:0005216
|
ion channel activity
|
0.369141 |
0.369141 |
1
|
189
|
|
GO:0000166
|
nucleotide binding
|
0.374077 |
0.374077 |
1
|
1178
|
|
GO:0006793
|
phosphorus metabolic process
|
0.376764 |
0.376764 |
1
|
556
|
|
GO:0044281
|
small molecule metabolic process
|
0.378352 |
0.378352 |
1
|
761
|
|
GO:0016491
|
oxidoreductase activity
|
0.397549 |
0.397549 |
1
|
638
|
|
GO:0044425
|
membrane part
|
0.400435 |
0.400435 |
1
|
1398
|
|
GO:0043085
|
positive regulation of catalytic activity
|
0.405063 |
0.405063 |
1
|
64
|
|
GO:0016070
|
RNA metabolic process
|
0.413829 |
0.413829 |
1
|
1191
|
|
GO:0022832
|
voltage-gated channel activity
|
0.415385 |
0.415385 |
1
|
54
|
|
GO:0023052
|
signaling
|
0.423150 |
0.423150 |
1
|
1364
|
|
GO:0032991
|
macromolecular complex
|
0.428425 |
0.428425 |
1
|
2151
|
|
GO:0023034
|
intracellular signaling pathway
|
0.430962 |
0.430962 |
1
|
412
|
|
GO:0044093
|
positive regulation of molecular function
|
0.437126 |
0.437126 |
1
|
73
|
|
GO:0051234
|
establishment of localization
|
0.467930 |
0.467930 |
1
|
1549
|
|
GO:0033674
|
positive regulation of kinase activity
|
0.471698 |
0.471698 |
1
|
25
|
|
GO:0045860
|
positive regulation of protein kinase activity
|
0.471698 |
0.471698 |
1
|
25
|
|
GO:0016772
|
transferase activity, transferring phosphorus-containing groups
|
0.480583 |
0.480583 |
1
|
495
|
|
GO:0008150
|
biological_process
|
0.481028 |
0.481028 |
4
|
10616
|
|
GO:0005623
|
cell
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.497519 |
0.497519 |
2
|
1959
|
|
GO:0019222
|
regulation of metabolic process
|
0.507779 |
0.507779 |
1
|
1242
|
|
GO:0010467
|
gene expression
|
0.531494 |
0.531494 |
1
|
1907
|
|
GO:0051179
|
localization
|
0.544835 |
0.544835 |
1
|
1896
|
|
GO:0003676
|
nucleic acid binding
|
0.549587 |
0.549587 |
1
|
1855
|
|
GO:0032147
|
activation of protein kinase activity
|
0.560000 |
0.560000 |
1
|
14
|
|
GO:0003674
|
molecular_function
|
0.567526 |
0.567526 |
4
|
11064
|
|
GO:0016020
|
membrane
|
0.567775 |
0.567775 |
1
|
2122
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.569226 |
0.569226 |
1
|
1110
|
|
GO:0016842
|
amidine-lyase activity
|
0.571429 |
0.571429 |
1
|
4
|
|
GO:0016740
|
transferase activity
|
0.579665 |
0.579665 |
1
|
1030
|
|
GO:0034703
|
cation channel complex
|
0.584906 |
0.584906 |
1
|
31
|
|
GO:0050789
|
regulation of biological process
|
0.613640 |
0.613640 |
1
|
2246
|
|
GO:0030001
|
metal ion transport
|
0.639676 |
0.639676 |
1
|
158
|
|
GO:0022836
|
gated channel activity
|
0.646766 |
0.646766 |
1
|
130
|
|
GO:0007166
|
cell surface receptor linked signaling pathway
|
0.652720 |
0.652720 |
1
|
624
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.658716 |
0.658716 |
1
|
1789
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0065007
|
biological regulation
|
0.666464 |
0.666464 |
1
|
2548
|
|
GO:0032502
|
developmental process
|
0.681102 |
0.681102 |
1
|
2638
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0005488
|
binding
|
0.702165 |
0.702165 |
2
|
5641
|
|
GO:0006399
|
tRNA metabolic process
|
0.705882 |
0.705882 |
1
|
84
|
|
GO:0031224
|
intrinsic to membrane
|
0.725322 |
0.725322 |
1
|
1014
|
|
GO:0043234
|
protein complex
|
0.727104 |
0.727104 |
1
|
1564
|
|
GO:0050662
|
coenzyme binding
|
0.727749 |
0.727749 |
1
|
139
|
|
GO:0016773
|
phosphotransferase activity, alcohol group as acceptor
|
0.735354 |
0.735354 |
1
|
364
|
|
GO:0006812
|
cation transport
|
0.746224 |
0.746224 |
1
|
247
|
|
GO:0042045
|
epithelial fluid transport
|
0.750000 |
0.750000 |
1
|
9
|
|
GO:0016310
|
phosphorylation
|
0.758993 |
0.758993 |
1
|
422
|
|
GO:0043549
|
regulation of kinase activity
|
0.768116 |
0.768116 |
1
|
53
|
|
GO:0050794
|
regulation of cellular process
|
0.768615 |
0.768615 |
1
|
2034
|
|
GO:0032501
|
multicellular organismal process
|
0.776347 |
0.776347 |
1
|
3315
|
|
GO:0009058
|
biosynthetic process
|
0.786651 |
0.786651 |
1
|
2090
|
|
GO:0022891
|
substrate-specific transmembrane transporter activity
|
0.788146 |
0.788146 |
1
|
625
|
|
GO:0016301
|
kinase activity
|
0.795960 |
0.795960 |
1
|
394
|
|
GO:0017076
|
purine nucleotide binding
|
0.803056 |
0.803056 |
1
|
946
|
|
GO:0008324
|
cation transmembrane transporter activity
|
0.808679 |
0.808679 |
1
|
410
|
|
GO:0015075
|
ion transmembrane transporter activity
|
0.811200 |
0.811200 |
1
|
507
|
|
GO:0022892
|
substrate-specific transporter activity
|
0.815728 |
0.815728 |
1
|
695
|
|
GO:0030554
|
adenyl nucleotide binding
|
0.819239 |
0.819239 |
1
|
775
|
|
GO:0022857
|
transmembrane transporter activity
|
0.852113 |
0.852113 |
1
|
726
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0006816
|
calcium ion transport
|
0.888889 |
0.888889 |
1
|
32
|
|
GO:0044249
|
cellular biosynthetic process
|
0.894138 |
0.894138 |
1
|
2034
|
|
GO:0005575
|
cellular_component
|
0.909916 |
0.909916 |
2
|
8817
|
|
GO:0044238
|
primary metabolic process
|
0.914505 |
0.914505 |
2
|
4259
|
|
GO:0043436
|
oxoacid metabolic process
|
0.925000 |
0.925000 |
1
|
259
|
|
GO:0005622
|
intracellular
|
0.944411 |
0.944411 |
1
|
4734
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.953060 |
0.953060 |
1
|
2878
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0043170
|
macromolecule metabolic process
|
0.970476 |
0.970476 |
1
|
3588
|
|
GO:0016021
|
integral to membrane
|
0.984221 |
0.984221 |
1
|
998
|
|
GO:0001883
|
purine nucleoside binding
|
0.992347 |
0.992347 |
1
|
778
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
4
|
12747
|
|
GO:0019752
|
carboxylic acid metabolic process
|
1.00000 |
1.00000 |
1
|
259
|
|
GO:0006936
|
muscle contraction
|
1.00000 |
1.00000 |
1
|
17
|
|
GO:0006796
|
phosphate metabolic process
|
1.00000 |
1.00000 |
1
|
556
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|
|
GO:0015267
|
channel activity
|
1.00000 |
1.00000 |
1
|
201
|
|
GO:0045859
|
regulation of protein kinase activity
|
1.00000 |
1.00000 |
1
|
53
|