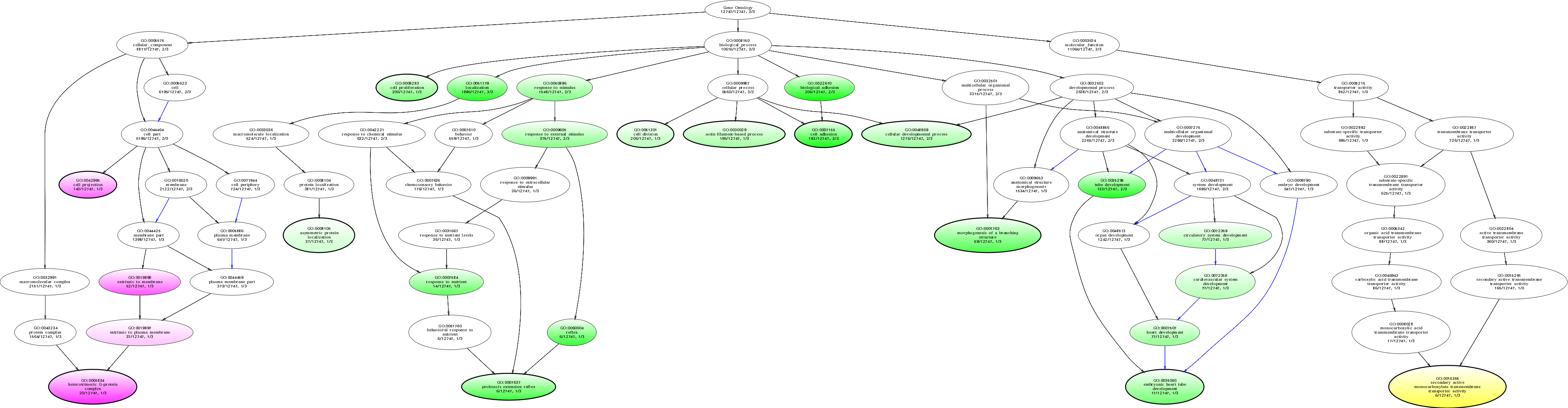
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0022610
|
biological adhesion
|
0.00110984 |
0.00110984 |
2
|
206
|
|
GO:0007155
|
cell adhesion
|
0.00249771 |
0.00249771 |
2
|
192
|
|
GO:0035295
|
tube development
|
0.00264053 |
0.00264053 |
2
|
133
|
|
GO:0051179
|
localization
|
0.00568941 |
0.00568941 |
3
|
1896
|
|
GO:0005834
|
heterotrimeric G-protein complex
|
0.0127389 |
0.0127389 |
1
|
20
|
|
GO:0060004
|
reflex
|
0.0317861 |
0.0317861 |
1
|
6
|
|
GO:0007637
|
proboscis extension reflex
|
0.0340909 |
0.0340909 |
1
|
6
|
|
GO:0015355
|
secondary active monocarboxylate transmembrane transporter activity
|
0.0346821 |
0.0346821 |
1
|
6
|
|
GO:0001763
|
morphogenesis of a branching structure
|
0.0347851 |
0.0347851 |
1
|
59
|
|
GO:0019898
|
extrinsic to membrane
|
0.0371960 |
0.0371960 |
1
|
52
|
|
GO:0007584
|
response to nutrient
|
0.0429727 |
0.0429727 |
1
|
14
|
|
GO:0042995
|
cell projection
|
0.0456371 |
0.0456371 |
1
|
143
|
|
GO:0035050
|
embryonic heart tube development
|
0.0466968 |
0.0466968 |
1
|
17
|
|
GO:0008283
|
cell proliferation
|
0.0554655 |
0.0554655 |
1
|
200
|
|
GO:0050896
|
response to stimulus
|
0.0575624 |
0.0575624 |
2
|
1548
|
|
GO:0009605
|
response to external stimulus
|
0.0585655 |
0.0585655 |
2
|
375
|
|
GO:0007507
|
heart development
|
0.0619968 |
0.0619968 |
1
|
77
|
|
GO:0048869
|
cellular developmental process
|
0.0826655 |
0.0826655 |
2
|
1276
|
|
GO:0030029
|
actin filament-based process
|
0.0826842 |
0.0826842 |
1
|
186
|
|
GO:0072359
|
circulatory system development
|
0.0887662 |
0.0887662 |
1
|
77
|
|
GO:0072358
|
cardiovascular system development
|
0.0887662 |
0.0887662 |
1
|
77
|
|
GO:0051301
|
cell division
|
0.0912934 |
0.0912934 |
1
|
206
|
|
GO:0019897
|
extrinsic to plasma membrane
|
0.0963542 |
0.0963542 |
1
|
37
|
|
GO:0008105
|
asymmetric protein localization
|
0.0971129 |
0.0971129 |
1
|
37
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.102602 |
0.102602 |
2
|
2108
|
|
GO:0016020
|
membrane
|
0.117293 |
0.117293 |
2
|
2122
|
|
GO:0055059
|
asymmetric neuroblast division
|
0.117647 |
0.117647 |
1
|
30
|
|
GO:0006928
|
cellular component movement
|
0.118060 |
0.118060 |
1
|
269
|
|
GO:0040011
|
locomotion
|
0.130645 |
0.130645 |
1
|
484
|
|
GO:0005342
|
organic acid transmembrane transporter activity
|
0.140800 |
0.140800 |
1
|
88
|
|
GO:0060089
|
molecular transducer activity
|
0.144056 |
0.144056 |
1
|
559
|
|
GO:0035239
|
tube morphogenesis
|
0.146222 |
0.146222 |
1
|
117
|
|
GO:0007405
|
neuroblast proliferation
|
0.146868 |
0.146868 |
1
|
46
|
|
GO:0008360
|
regulation of cell shape
|
0.153635 |
0.153635 |
1
|
112
|
|
GO:0032502
|
developmental process
|
0.154531 |
0.154531 |
2
|
2638
|
|
GO:0035146
|
tube fusion
|
0.162393 |
0.162393 |
1
|
19
|
|
GO:0042221
|
response to chemical stimulus
|
0.166527 |
0.166527 |
2
|
632
|
|
GO:0009991
|
response to extracellular stimulus
|
0.183016 |
0.183016 |
1
|
36
|
|
GO:0055085
|
transmembrane transport
|
0.191633 |
0.191633 |
1
|
469
|
|
GO:0061138
|
morphogenesis of a branching epithelium
|
0.192440 |
0.192440 |
1
|
56
|
|
GO:0008045
|
motor axon guidance
|
0.193548 |
0.193548 |
1
|
36
|
|
GO:0048729
|
tissue morphogenesis
|
0.194151 |
0.194151 |
1
|
312
|
|
GO:0008028
|
monocarboxylic acid transmembrane transporter activity
|
0.197674 |
0.197674 |
1
|
17
|
|
GO:0022008
|
neurogenesis
|
0.204486 |
0.204486 |
2
|
632
|
|
GO:0030030
|
cell projection organization
|
0.205631 |
0.205631 |
1
|
485
|
|
GO:0060446
|
branching involved in open tracheal system development
|
0.208178 |
0.208178 |
1
|
56
|
|
GO:0005215
|
transporter activity
|
0.213704 |
0.213704 |
1
|
852
|
|
GO:0071944
|
cell periphery
|
0.220095 |
0.220095 |
1
|
724
|
|
GO:0061351
|
neural precursor cell proliferation
|
0.230000 |
0.230000 |
1
|
46
|
|
GO:0032501
|
multicellular organismal process
|
0.231607 |
0.231607 |
2
|
3315
|
|
GO:0009987
|
cellular process
|
0.235914 |
0.235914 |
3
|
6560
|
|
GO:0044459
|
plasma membrane part
|
0.236675 |
0.236675 |
1
|
373
|
|
GO:0003924
|
GTPase activity
|
0.241796 |
0.241796 |
1
|
140
|
|
GO:0045176
|
apical protein localization
|
0.243243 |
0.243243 |
1
|
9
|
|
GO:0007399
|
nervous system development
|
0.249853 |
0.249853 |
2
|
848
|
|
GO:0060541
|
respiratory system development
|
0.272151 |
0.272151 |
1
|
249
|
|
GO:0060562
|
epithelial tube morphogenesis
|
0.281150 |
0.281150 |
1
|
88
|
|
GO:0065008
|
regulation of biological quality
|
0.286107 |
0.286107 |
1
|
729
|
|
GO:0016817
|
hydrolase activity, acting on acid anhydrides
|
0.301217 |
0.301217 |
1
|
594
|
|
GO:0005515
|
protein binding
|
0.305974 |
0.305974 |
1
|
1726
|
|
GO:0007186
|
G-protein coupled receptor protein signaling pathway
|
0.318910 |
0.318910 |
1
|
199
|
|
GO:0023052
|
signaling
|
0.338079 |
0.338079 |
1
|
1364
|
|
GO:0007635
|
chemosensory behavior
|
0.338282 |
0.338282 |
1
|
176
|
|
GO:0035147
|
branch fusion, open tracheal system
|
0.339286 |
0.339286 |
1
|
19
|
|
GO:0030036
|
actin cytoskeleton organization
|
0.339518 |
0.339518 |
1
|
183
|
|
GO:0017145
|
stem cell division
|
0.339806 |
0.339806 |
1
|
70
|
|
GO:0032989
|
cellular component morphogenesis
|
0.348979 |
0.348979 |
1
|
566
|
|
GO:0045165
|
cell fate commitment
|
0.359875 |
0.359875 |
1
|
255
|
|
GO:0051674
|
localization of cell
|
0.361247 |
0.361247 |
1
|
263
|
|
GO:0000904
|
cell morphogenesis involved in differentiation
|
0.376731 |
0.376731 |
1
|
408
|
|
GO:0051234
|
establishment of localization
|
0.377001 |
0.377001 |
1
|
1549
|
|
GO:0044425
|
membrane part
|
0.400435 |
0.400435 |
1
|
1398
|
|
GO:0009790
|
embryo development
|
0.427001 |
0.427001 |
1
|
641
|
|
GO:0032991
|
macromolecular complex
|
0.428425 |
0.428425 |
1
|
2151
|
|
GO:0051780
|
behavioral response to nutrient
|
0.428571 |
0.428571 |
1
|
6
|
|
GO:0009888
|
tissue development
|
0.431439 |
0.431439 |
1
|
557
|
|
GO:0048731
|
system development
|
0.432376 |
0.432376 |
2
|
1696
|
|
GO:0048666
|
neuron development
|
0.433702 |
0.433702 |
1
|
471
|
|
GO:0007275
|
multicellular organismal development
|
0.442066 |
0.442066 |
2
|
2296
|
|
GO:0007010
|
cytoskeleton organization
|
0.453469 |
0.453469 |
1
|
536
|
|
GO:0015291
|
secondary active transmembrane transporter activity
|
0.458333 |
0.458333 |
1
|
165
|
|
GO:0048754
|
branching morphogenesis of a tube
|
0.466667 |
0.466667 |
1
|
56
|
|
GO:0016787
|
hydrolase activity
|
0.480273 |
0.480273 |
1
|
1972
|
|
GO:0006935
|
chemotaxis
|
0.492835 |
0.492835 |
1
|
193
|
|
GO:0005623
|
cell
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0022804
|
active transmembrane transporter activity
|
0.495868 |
0.495868 |
1
|
360
|
|
GO:0005886
|
plasma membrane
|
0.498869 |
0.498869 |
1
|
641
|
|
GO:0048870
|
cell motility
|
0.512097 |
0.512097 |
1
|
254
|
|
GO:0060429
|
epithelium development
|
0.529623 |
0.529623 |
1
|
295
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.556542 |
0.556542 |
1
|
1562
|
|
GO:0048103
|
somatic stem cell division
|
0.557143 |
0.557143 |
1
|
39
|
|
GO:0065007
|
biological regulation
|
0.561089 |
0.561089 |
1
|
2548
|
|
GO:0055057
|
neuroblast division
|
0.574074 |
0.574074 |
1
|
31
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0007610
|
behavior
|
0.591970 |
0.591970 |
1
|
559
|
|
GO:0007015
|
actin filament organization
|
0.606557 |
0.606557 |
1
|
111
|
|
GO:0033036
|
macromolecule localization
|
0.621309 |
0.621309 |
1
|
524
|
|
GO:0042330
|
taxis
|
0.628051 |
0.628051 |
1
|
237
|
|
GO:0007166
|
cell surface receptor linked signaling pathway
|
0.652720 |
0.652720 |
1
|
624
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0007409
|
axonogenesis
|
0.679389 |
0.679389 |
1
|
267
|
|
GO:0007411
|
axon guidance
|
0.683824 |
0.683824 |
1
|
186
|
|
GO:0030182
|
neuron differentiation
|
0.689161 |
0.689161 |
1
|
548
|
|
GO:0031175
|
neuron projection development
|
0.696270 |
0.696270 |
1
|
392
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0048468
|
cell development
|
0.705715 |
0.705715 |
1
|
1042
|
|
GO:0008104
|
protein localization
|
0.727099 |
0.727099 |
1
|
381
|
|
GO:0043234
|
protein complex
|
0.727104 |
0.727104 |
1
|
1564
|
|
GO:0048856
|
anatomical structure development
|
0.737157 |
0.737157 |
2
|
2265
|
|
GO:0032990
|
cell part morphogenesis
|
0.745583 |
0.745583 |
1
|
422
|
|
GO:0003824
|
catalytic activity
|
0.751316 |
0.751316 |
1
|
4106
|
|
GO:0005575
|
cellular_component
|
0.773470 |
0.773470 |
2
|
8817
|
|
GO:0006996
|
organelle organization
|
0.779169 |
0.779169 |
1
|
1182
|
|
GO:0022891
|
substrate-specific transmembrane transporter activity
|
0.788146 |
0.788146 |
1
|
625
|
|
GO:0048667
|
cell morphogenesis involved in neuron differentiation
|
0.795501 |
0.795501 |
1
|
389
|
|
GO:0048513
|
organ development
|
0.796117 |
0.796117 |
1
|
1242
|
|
GO:0048858
|
cell projection morphogenesis
|
0.806883 |
0.806883 |
1
|
422
|
|
GO:0022892
|
substrate-specific transporter activity
|
0.815728 |
0.815728 |
1
|
695
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.824951 |
0.824951 |
1
|
1534
|
|
GO:0000902
|
cell morphogenesis
|
0.825088 |
0.825088 |
1
|
467
|
|
GO:0022857
|
transmembrane transporter activity
|
0.852113 |
0.852113 |
1
|
726
|
|
GO:0005488
|
binding
|
0.882277 |
0.882277 |
1
|
5641
|
|
GO:0002009
|
morphogenesis of an epithelium
|
0.908805 |
0.908805 |
1
|
289
|
|
GO:0048699
|
generation of neurons
|
0.910264 |
0.910264 |
2
|
603
|
|
GO:0048812
|
neuron projection morphogenesis
|
0.910798 |
0.910798 |
1
|
388
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.923977 |
0.923977 |
1
|
1517
|
|
GO:0030154
|
cell differentiation
|
0.942825 |
0.942825 |
2
|
1239
|
|
GO:0016043
|
cellular component organization
|
0.947563 |
0.947563 |
2
|
2052
|
|
GO:0016477
|
cell migration
|
0.964567 |
0.964567 |
1
|
245
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0046943
|
carboxylic acid transmembrane transporter activity
|
0.977273 |
0.977273 |
1
|
86
|
|
GO:0017111
|
nucleoside-triphosphatase activity
|
0.983022 |
0.983022 |
1
|
579
|
|
GO:0016462
|
pyrophosphatase activity
|
0.991582 |
0.991582 |
1
|
589
|
|
GO:0007424
|
open tracheal system development
|
0.995984 |
0.995984 |
1
|
248
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0016818
|
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
|
1.00000 |
1.00000 |
1
|
594
|
|
GO:0004871
|
signal transducer activity
|
1.00000 |
1.00000 |
1
|
559
|
|
GO:0031667
|
response to nutrient levels
|
1.00000 |
1.00000 |
1
|
36
|