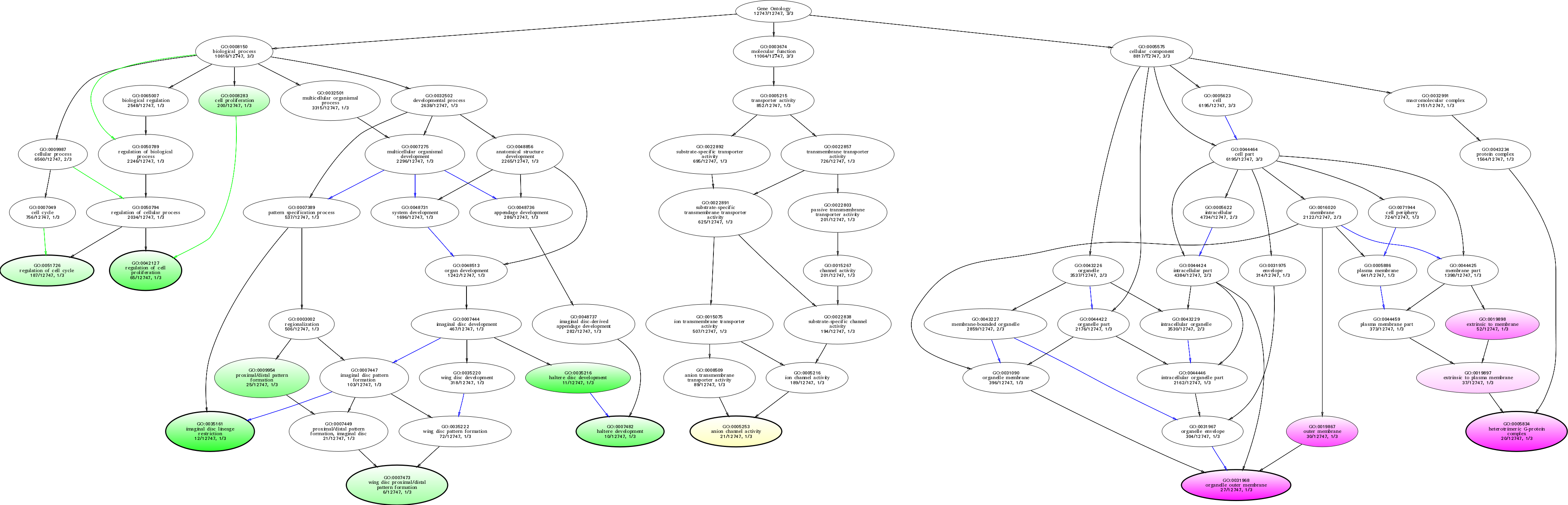
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0031968
|
organelle outer membrane
|
0.0122698 |
0.0122698 |
1
|
27
|
|
GO:0005834
|
heterotrimeric G-protein complex
|
0.0127389 |
0.0127389 |
1
|
20
|
|
GO:0035161
|
imaginal disc lineage restriction
|
0.0223464 |
0.0223464 |
1
|
12
|
|
GO:0035216
|
haltere disc development
|
0.0235546 |
0.0235546 |
1
|
11
|
|
GO:0019867
|
outer membrane
|
0.0280819 |
0.0280819 |
1
|
30
|
|
GO:0042127
|
regulation of cell proliferation
|
0.0311005 |
0.0311005 |
1
|
65
|
|
GO:0007482
|
haltere development
|
0.0354610 |
0.0354610 |
1
|
10
|
|
GO:0019898
|
extrinsic to membrane
|
0.0371960 |
0.0371960 |
1
|
52
|
|
GO:0009954
|
proximal/distal pattern formation
|
0.0494071 |
0.0494071 |
1
|
25
|
|
GO:0008283
|
cell proliferation
|
0.0554655 |
0.0554655 |
1
|
200
|
|
GO:0007473
|
wing disc proximal/distal pattern formation
|
0.0705882 |
0.0705882 |
1
|
6
|
|
GO:0051726
|
regulation of cell cycle
|
0.0752212 |
0.0752212 |
1
|
187
|
|
GO:0005253
|
anion channel activity
|
0.0817121 |
0.0817121 |
1
|
21
|
|
GO:0019897
|
extrinsic to plasma membrane
|
0.0963542 |
0.0963542 |
1
|
37
|
|
GO:0048736
|
appendage development
|
0.110896 |
0.110896 |
1
|
286
|
|
GO:0061061
|
muscle structure development
|
0.111258 |
0.111258 |
1
|
252
|
|
GO:0007478
|
leg disc morphogenesis
|
0.111782 |
0.111782 |
1
|
37
|
|
GO:0030528
|
transcription regulator activity
|
0.112632 |
0.112632 |
1
|
432
|
|
GO:0005741
|
mitochondrial outer membrane
|
0.120370 |
0.120370 |
1
|
26
|
|
GO:0035218
|
leg disc development
|
0.122056 |
0.122056 |
1
|
57
|
|
GO:0044429
|
mitochondrial part
|
0.125285 |
0.125285 |
1
|
384
|
|
GO:0035127
|
post-embryonic limb morphogenesis
|
0.128205 |
0.128205 |
1
|
35
|
|
GO:0035109
|
imaginal disc-derived limb morphogenesis
|
0.132143 |
0.132143 |
1
|
37
|
|
GO:0055085
|
transmembrane transport
|
0.132219 |
0.132219 |
1
|
469
|
|
GO:0007447
|
imaginal disc pattern formation
|
0.133075 |
0.133075 |
1
|
103
|
|
GO:0007451
|
dorsal/ventral lineage restriction, imaginal disc
|
0.133333 |
0.133333 |
1
|
8
|
|
GO:0006820
|
anion transport
|
0.141994 |
0.141994 |
1
|
47
|
|
GO:0031975
|
envelope
|
0.144503 |
0.144503 |
1
|
314
|
|
GO:0008308
|
voltage-gated anion channel activity
|
0.153846 |
0.153846 |
1
|
10
|
|
GO:0008587
|
imaginal disc-derived wing margin morphogenesis
|
0.155340 |
0.155340 |
1
|
64
|
|
GO:0060173
|
limb development
|
0.164336 |
0.164336 |
1
|
47
|
|
GO:0035108
|
limb morphogenesis
|
0.166078 |
0.166078 |
1
|
47
|
|
GO:0007525
|
somatic muscle development
|
0.166667 |
0.166667 |
1
|
42
|
|
GO:0031967
|
organelle envelope
|
0.169005 |
0.169005 |
1
|
304
|
|
GO:0008509
|
anion transmembrane transporter activity
|
0.175542 |
0.175542 |
1
|
89
|
|
GO:0035107
|
appendage morphogenesis
|
0.184365 |
0.184365 |
1
|
283
|
|
GO:0009791
|
post-embryonic development
|
0.189538 |
0.189538 |
1
|
500
|
|
GO:0007449
|
proximal/distal pattern formation, imaginal disc
|
0.196262 |
0.196262 |
1
|
21
|
|
GO:0007389
|
pattern specification process
|
0.203563 |
0.203563 |
1
|
537
|
|
GO:0019222
|
regulation of metabolic process
|
0.210401 |
0.210401 |
1
|
1242
|
|
GO:0035222
|
wing disc pattern formation
|
0.212389 |
0.212389 |
1
|
72
|
|
GO:0005215
|
transporter activity
|
0.213704 |
0.213704 |
1
|
852
|
|
GO:0007049
|
cell cycle
|
0.217222 |
0.217222 |
1
|
756
|
|
GO:0007346
|
regulation of mitotic cell cycle
|
0.220165 |
0.220165 |
1
|
107
|
|
GO:0006811
|
ion transport
|
0.221109 |
0.221109 |
1
|
331
|
|
GO:0031090
|
organelle membrane
|
0.221394 |
0.221394 |
1
|
396
|
|
GO:0044459
|
plasma membrane part
|
0.236675 |
0.236675 |
1
|
373
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.237215 |
0.237215 |
1
|
1039
|
|
GO:0003924
|
GTPase activity
|
0.241796 |
0.241796 |
1
|
140
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.244709 |
0.244709 |
1
|
1110
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.259446 |
0.259446 |
1
|
412
|
|
GO:0016020
|
membrane
|
0.271576 |
0.271576 |
2
|
2122
|
|
GO:0007552
|
metamorphosis
|
0.273794 |
0.273794 |
1
|
420
|
|
GO:0048569
|
post-embryonic organ development
|
0.276167 |
0.276167 |
1
|
343
|
|
GO:0022803
|
passive transmembrane transporter activity
|
0.276860 |
0.276860 |
1
|
201
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.283034 |
0.283034 |
1
|
1056
|
|
GO:0005739
|
mitochondrion
|
0.284169 |
0.284169 |
1
|
551
|
|
GO:0005244
|
voltage-gated ion channel activity
|
0.285714 |
0.285714 |
1
|
54
|
|
GO:0009953
|
dorsal/ventral pattern formation
|
0.286561 |
0.286561 |
1
|
145
|
|
GO:0007450
|
dorsal/ventral pattern formation, imaginal disc
|
0.296296 |
0.296296 |
1
|
56
|
|
GO:0016817
|
hydrolase activity, acting on acid anhydrides
|
0.301217 |
0.301217 |
1
|
594
|
|
GO:0022838
|
substrate-specific channel activity
|
0.307448 |
0.307448 |
1
|
194
|
|
GO:0071944
|
cell periphery
|
0.311271 |
0.311271 |
1
|
724
|
|
GO:0007186
|
G-protein coupled receptor protein signaling pathway
|
0.318910 |
0.318910 |
1
|
199
|
|
GO:0005575
|
cellular_component
|
0.330897 |
0.330897 |
3
|
8817
|
|
GO:0023052
|
signaling
|
0.338079 |
0.338079 |
1
|
1364
|
|
GO:0005623
|
cell
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0044464
|
cell part
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0043226
|
organelle
|
0.353651 |
0.353651 |
2
|
3537
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.366791 |
0.366791 |
1
|
888
|
|
GO:0005216
|
ion channel activity
|
0.369141 |
0.369141 |
1
|
189
|
|
GO:0009887
|
organ morphogenesis
|
0.374384 |
0.374384 |
1
|
684
|
|
GO:0007444
|
imaginal disc development
|
0.376006 |
0.376006 |
1
|
467
|
|
GO:0051234
|
establishment of localization
|
0.377001 |
0.377001 |
1
|
1549
|
|
GO:0031966
|
mitochondrial membrane
|
0.380531 |
0.380531 |
1
|
215
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.381695 |
0.381695 |
1
|
905
|
|
GO:0006350
|
transcription
|
0.387810 |
0.387810 |
1
|
859
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.396252 |
0.396252 |
1
|
888
|
|
GO:0009058
|
biosynthetic process
|
0.402387 |
0.402387 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.402965 |
0.402965 |
1
|
2093
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.405649 |
0.405649 |
1
|
1623
|
|
GO:0048563
|
post-embryonic organ morphogenesis
|
0.415167 |
0.415167 |
1
|
323
|
|
GO:0022832
|
voltage-gated channel activity
|
0.415385 |
0.415385 |
1
|
54
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.415756 |
0.415756 |
1
|
1789
|
|
GO:0007423
|
sensory organ development
|
0.438003 |
0.438003 |
1
|
544
|
|
GO:0051179
|
localization
|
0.445834 |
0.445834 |
1
|
1896
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.451726 |
0.451726 |
1
|
903
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.456989 |
0.456989 |
1
|
850
|
|
GO:0016787
|
hydrolase activity
|
0.480273 |
0.480273 |
1
|
1972
|
|
GO:0010468
|
regulation of gene expression
|
0.489930 |
0.489930 |
1
|
973
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.496927 |
0.496927 |
1
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.498346 |
0.498346 |
1
|
1959
|
|
GO:0005886
|
plasma membrane
|
0.498869 |
0.498869 |
1
|
641
|
|
GO:0050789
|
regulation of biological process
|
0.509927 |
0.509927 |
1
|
2246
|
|
GO:0005740
|
mitochondrial envelope
|
0.512088 |
0.512088 |
1
|
233
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.513595 |
0.513595 |
1
|
850
|
|
GO:0050794
|
regulation of cellular process
|
0.518911 |
0.518911 |
1
|
2034
|
|
GO:0044249
|
cellular biosynthetic process
|
0.526807 |
0.526807 |
1
|
2034
|
|
GO:0010467
|
gene expression
|
0.531494 |
0.531494 |
1
|
1907
|
|
GO:0044425
|
membrane part
|
0.535780 |
0.535780 |
1
|
1398
|
|
GO:0048592
|
eye morphogenesis
|
0.542069 |
0.542069 |
1
|
393
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0065007
|
biological regulation
|
0.561089 |
0.561089 |
1
|
2548
|
|
GO:0000278
|
mitotic cell cycle
|
0.567460 |
0.567460 |
1
|
429
|
|
GO:0032991
|
macromolecular complex
|
0.567899 |
0.567899 |
1
|
2151
|
|
GO:0044422
|
organelle part
|
0.572743 |
0.572743 |
1
|
2176
|
|
GO:0032502
|
developmental process
|
0.575616 |
0.575616 |
1
|
2638
|
|
GO:0007560
|
imaginal disc morphogenesis
|
0.575758 |
0.575758 |
1
|
323
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0048190
|
wing disc dorsal/ventral pattern formation
|
0.636364 |
0.636364 |
1
|
49
|
|
GO:0035120
|
post-embryonic appendage morphogenesis
|
0.638095 |
0.638095 |
1
|
268
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.639129 |
0.639129 |
1
|
2878
|
|
GO:0043229
|
intracellular organelle
|
0.646541 |
0.646541 |
2
|
3530
|
|
GO:0022836
|
gated channel activity
|
0.646766 |
0.646766 |
1
|
130
|
|
GO:0007166
|
cell surface receptor linked signaling pathway
|
0.652720 |
0.652720 |
1
|
624
|
|
GO:0043227
|
membrane-bounded organelle
|
0.653325 |
0.653325 |
2
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.653801 |
0.653801 |
2
|
2856
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0007472
|
wing disc morphogenesis
|
0.665796 |
0.665796 |
1
|
255
|
|
GO:0044444
|
cytoplasmic part
|
0.669378 |
0.669378 |
1
|
1863
|
|
GO:0009987
|
cellular process
|
0.673638 |
0.673638 |
2
|
6560
|
|
GO:0032501
|
multicellular organismal process
|
0.674757 |
0.674757 |
1
|
3315
|
|
GO:0035220
|
wing disc development
|
0.680942 |
0.680942 |
1
|
318
|
|
GO:0045449
|
regulation of transcription
|
0.687166 |
0.687166 |
1
|
771
|
|
GO:0043170
|
macromolecule metabolic process
|
0.690797 |
0.690797 |
1
|
3588
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0043234
|
protein complex
|
0.727104 |
0.727104 |
1
|
1564
|
|
GO:0007480
|
imaginal disc-derived leg morphogenesis
|
0.729167 |
0.729167 |
1
|
35
|
|
GO:0044237
|
cellular metabolic process
|
0.740940 |
0.740940 |
1
|
3814
|
|
GO:0044446
|
intracellular organelle part
|
0.743053 |
0.743053 |
1
|
2162
|
|
GO:0003824
|
catalytic activity
|
0.751316 |
0.751316 |
1
|
4106
|
|
GO:0022891
|
substrate-specific transmembrane transporter activity
|
0.788146 |
0.788146 |
1
|
625
|
|
GO:0005737
|
cytoplasm
|
0.790684 |
0.790684 |
1
|
2378
|
|
GO:0044424
|
intracellular part
|
0.793631 |
0.793631 |
2
|
4384
|
|
GO:0015075
|
ion transmembrane transporter activity
|
0.811200 |
0.811200 |
1
|
507
|
|
GO:0001745
|
compound eye morphogenesis
|
0.811947 |
0.811947 |
1
|
367
|
|
GO:0022892
|
substrate-specific transporter activity
|
0.815728 |
0.815728 |
1
|
695
|
|
GO:0044238
|
primary metabolic process
|
0.819985 |
0.819985 |
1
|
4259
|
|
GO:0001654
|
eye development
|
0.841912 |
0.841912 |
1
|
458
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.842767 |
0.842767 |
1
|
402
|
|
GO:0022857
|
transmembrane transporter activity
|
0.852113 |
0.852113 |
1
|
726
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0005622
|
intracellular
|
0.859425 |
0.859425 |
2
|
4734
|
|
GO:0008152
|
metabolic process
|
0.866808 |
0.866808 |
1
|
5194
|
|
GO:0005634
|
nucleus
|
0.870016 |
0.870016 |
1
|
1826
|
|
GO:0007476
|
imaginal disc-derived wing morphogenesis
|
0.903915 |
0.903915 |
1
|
254
|
|
GO:0048749
|
compound eye development
|
0.932314 |
0.932314 |
1
|
427
|
|
GO:0003002
|
regionalization
|
0.942272 |
0.942272 |
1
|
506
|
|
GO:0002165
|
instar larval or pupal development
|
0.954000 |
0.954000 |
1
|
477
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0035114
|
imaginal disc-derived appendage morphogenesis
|
0.975524 |
0.975524 |
1
|
279
|
|
GO:0035110
|
leg morphogenesis
|
0.978723 |
0.978723 |
1
|
46
|
|
GO:0017111
|
nucleoside-triphosphatase activity
|
0.983022 |
0.983022 |
1
|
579
|
|
GO:0048737
|
imaginal disc-derived appendage development
|
0.986014 |
0.986014 |
1
|
282
|
|
GO:0016462
|
pyrophosphatase activity
|
0.991582 |
0.991582 |
1
|
589
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0016818
|
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
|
1.00000 |
1.00000 |
1
|
594
|
|
GO:0015267
|
channel activity
|
1.00000 |
1.00000 |
1
|
201
|