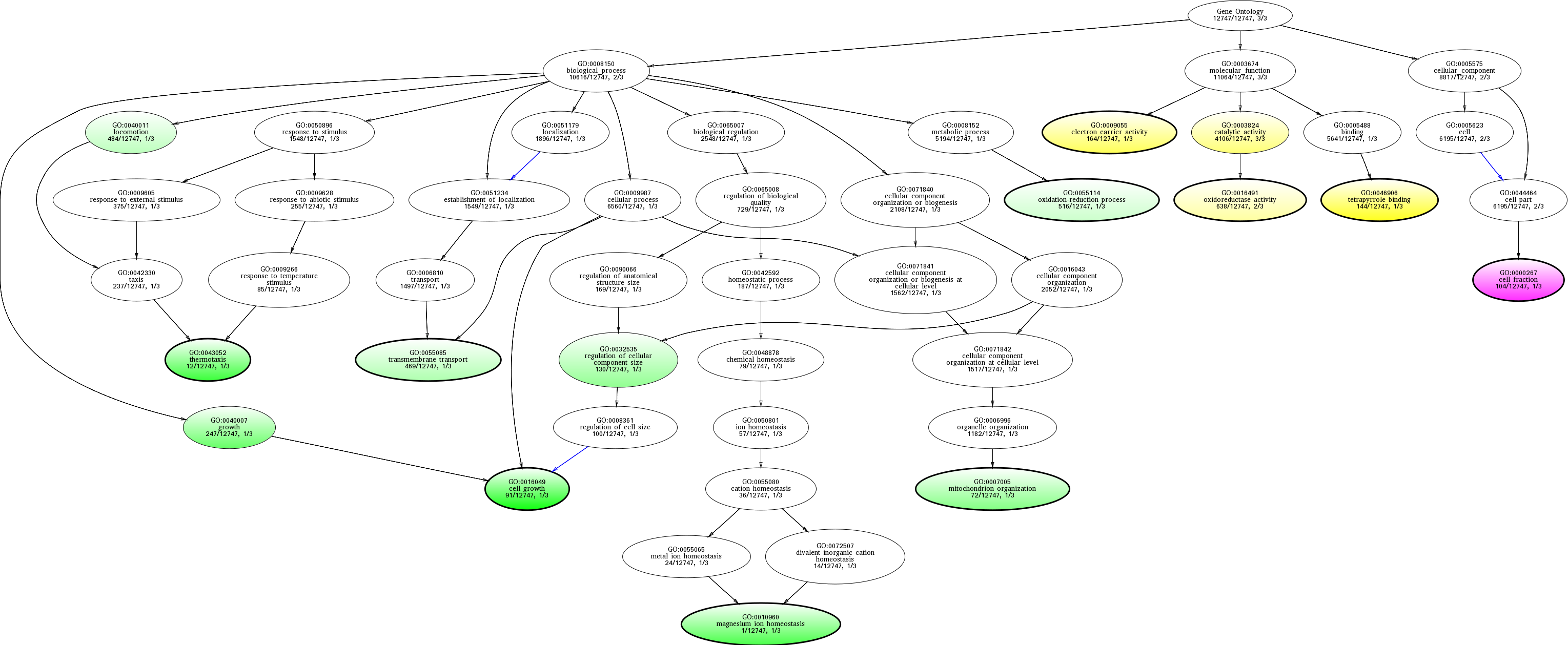
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0016049
|
cell growth
|
0.0138277 |
0.0138277 |
1
|
91
|
|
GO:0046906
|
tetrapyrrole binding
|
0.0255274 |
0.0255274 |
1
|
144
|
|
GO:0000267
|
cell fraction
|
0.0332963 |
0.0332963 |
1
|
104
|
|
GO:0043052
|
thermotaxis
|
0.0389610 |
0.0389610 |
1
|
12
|
|
GO:0010960
|
magnesium ion homeostasis
|
0.0400000 |
0.0400000 |
1
|
1
|
|
GO:0009055
|
electron carrier activity
|
0.0438166 |
0.0438166 |
1
|
164
|
|
GO:0040007
|
growth
|
0.0459943 |
0.0459943 |
1
|
247
|
|
GO:0003824
|
catalytic activity
|
0.0510882 |
0.0510882 |
3
|
4106
|
|
GO:0007005
|
mitochondrion organization
|
0.0609137 |
0.0609137 |
1
|
72
|
|
GO:0032535
|
regulation of cellular component size
|
0.0631681 |
0.0631681 |
1
|
130
|
|
GO:0016491
|
oxidoreductase activity
|
0.0648619 |
0.0648619 |
2
|
638
|
|
GO:0055085
|
transmembrane transport
|
0.0684472 |
0.0684472 |
1
|
469
|
|
GO:0040011
|
locomotion
|
0.0891086 |
0.0891086 |
1
|
484
|
|
GO:0055114
|
oxidation-reduction process
|
0.0993454 |
0.0993454 |
1
|
516
|
|
GO:0016020
|
membrane
|
0.117293 |
0.117293 |
2
|
2122
|
|
GO:0055069
|
zinc ion homeostasis
|
0.142857 |
0.142857 |
1
|
2
|
|
GO:0072511
|
divalent inorganic cation transport
|
0.149798 |
0.149798 |
1
|
37
|
|
GO:0009628
|
response to abiotic stimulus
|
0.164729 |
0.164729 |
1
|
255
|
|
GO:0005506
|
iron ion binding
|
0.177797 |
0.177797 |
1
|
205
|
|
GO:0005215
|
transporter activity
|
0.213704 |
0.213704 |
1
|
852
|
|
GO:0006811
|
ion transport
|
0.221109 |
0.221109 |
1
|
331
|
|
GO:0070838
|
divalent metal ion transport
|
0.227848 |
0.227848 |
1
|
36
|
|
GO:0090066
|
regulation of anatomical structure size
|
0.231824 |
0.231824 |
1
|
169
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.237386 |
0.237386 |
1
|
1562
|
|
GO:0009605
|
response to external stimulus
|
0.242248 |
0.242248 |
1
|
375
|
|
GO:0042592
|
homeostatic process
|
0.256516 |
0.256516 |
1
|
187
|
|
GO:0043167
|
ion binding
|
0.265556 |
0.265556 |
1
|
1498
|
|
GO:0046873
|
metal ion transmembrane transporter activity
|
0.268293 |
0.268293 |
1
|
110
|
|
GO:0050896
|
response to stimulus
|
0.270384 |
0.270384 |
1
|
1548
|
|
GO:0051234
|
establishment of localization
|
0.270545 |
0.270545 |
1
|
1549
|
|
GO:0022803
|
passive transmembrane transporter activity
|
0.276860 |
0.276860 |
1
|
201
|
|
GO:0065008
|
regulation of biological quality
|
0.286107 |
0.286107 |
1
|
729
|
|
GO:0022838
|
substrate-specific channel activity
|
0.307448 |
0.307448 |
1
|
194
|
|
GO:0051179
|
localization
|
0.325313 |
0.325313 |
1
|
1896
|
|
GO:0004497
|
monooxygenase activity
|
0.325669 |
0.325669 |
1
|
114
|
|
GO:0009266
|
response to temperature stimulus
|
0.333333 |
0.333333 |
1
|
85
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.357722 |
0.357722 |
1
|
2108
|
|
GO:0005216
|
ion channel activity
|
0.369141 |
0.369141 |
1
|
189
|
|
GO:0072507
|
divalent inorganic cation homeostasis
|
0.388889 |
0.388889 |
1
|
14
|
|
GO:0042330
|
taxis
|
0.389803 |
0.389803 |
1
|
237
|
|
GO:0065007
|
biological regulation
|
0.422440 |
0.422440 |
1
|
2548
|
|
GO:0048878
|
chemical homeostasis
|
0.422460 |
0.422460 |
1
|
79
|
|
GO:0032502
|
developmental process
|
0.435255 |
0.435255 |
1
|
2638
|
|
GO:0007423
|
sensory organ development
|
0.438003 |
0.438003 |
1
|
544
|
|
GO:0016772
|
transferase activity, transferring phosphorus-containing groups
|
0.480583 |
0.480583 |
1
|
495
|
|
GO:0005623
|
cell
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0032501
|
multicellular organismal process
|
0.527040 |
0.527040 |
1
|
3315
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0016740
|
transferase activity
|
0.579665 |
0.579665 |
1
|
1030
|
|
GO:0055080
|
cation homeostasis
|
0.631579 |
0.631579 |
1
|
36
|
|
GO:0030001
|
metal ion transport
|
0.639676 |
0.639676 |
1
|
158
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0055065
|
metal ion homeostasis
|
0.666667 |
0.666667 |
1
|
24
|
|
GO:0020037
|
heme binding
|
0.694175 |
0.694175 |
1
|
143
|
|
GO:0050801
|
ion homeostasis
|
0.721519 |
0.721519 |
1
|
57
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.724105 |
0.724105 |
1
|
1517
|
|
GO:0004672
|
protein kinase activity
|
0.728643 |
0.728643 |
1
|
290
|
|
GO:0016773
|
phosphotransferase activity, alcohol group as acceptor
|
0.735354 |
0.735354 |
1
|
364
|
|
GO:0008152
|
metabolic process
|
0.739170 |
0.739170 |
1
|
5194
|
|
GO:0006812
|
cation transport
|
0.746224 |
0.746224 |
1
|
247
|
|
GO:0008361
|
regulation of cell size
|
0.769231 |
0.769231 |
1
|
100
|
|
GO:0005575
|
cellular_component
|
0.773470 |
0.773470 |
2
|
8817
|
|
GO:0006996
|
organelle organization
|
0.779169 |
0.779169 |
1
|
1182
|
|
GO:0022891
|
substrate-specific transmembrane transporter activity
|
0.788146 |
0.788146 |
1
|
625
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0016301
|
kinase activity
|
0.795960 |
0.795960 |
1
|
394
|
|
GO:0008324
|
cation transmembrane transporter activity
|
0.808679 |
0.808679 |
1
|
410
|
|
GO:0015075
|
ion transmembrane transporter activity
|
0.811200 |
0.811200 |
1
|
507
|
|
GO:0022892
|
substrate-specific transporter activity
|
0.815728 |
0.815728 |
1
|
695
|
|
GO:0022857
|
transmembrane transporter activity
|
0.852113 |
0.852113 |
1
|
726
|
|
GO:0009987
|
cellular process
|
0.854049 |
0.854049 |
1
|
6560
|
|
GO:0042598
|
vesicular fraction
|
0.858586 |
0.858586 |
1
|
85
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0005488
|
binding
|
0.882277 |
0.882277 |
1
|
5641
|
|
GO:0008150
|
biological_process
|
0.925522 |
0.925522 |
2
|
10616
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0005624
|
membrane fraction
|
0.970588 |
0.970588 |
1
|
99
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0005626
|
insoluble fraction
|
0.980769 |
0.980769 |
1
|
102
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0005792
|
microsome
|
1.00000 |
1.00000 |
1
|
85
|
|
GO:0015267
|
channel activity
|
1.00000 |
1.00000 |
1
|
201
|