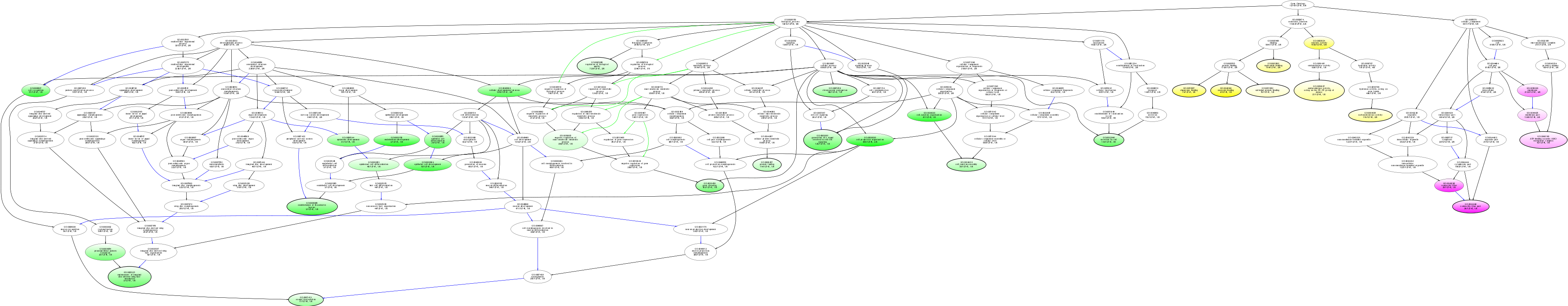
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0042043
|
neurexin binding
|
0.00463096 |
0.00463096 |
1
|
4
|
|
GO:0030030
|
cell projection organization
|
0.00544401 |
0.00544401 |
2
|
485
|
|
GO:0044449
|
contractile fiber part
|
0.00848010 |
0.00848010 |
1
|
26
|
|
GO:0003158
|
endothelium development
|
0.0101695 |
0.0101695 |
1
|
3
|
|
GO:0043292
|
contractile fiber
|
0.0107157 |
0.0107157 |
1
|
28
|
|
GO:0034330
|
cell junction organization
|
0.0142639 |
0.0142639 |
1
|
47
|
|
GO:0008037
|
cell recognition
|
0.0147896 |
0.0147896 |
1
|
57
|
|
GO:0051087
|
chaperone binding
|
0.0161614 |
0.0161614 |
1
|
14
|
|
GO:0002064
|
epithelial cell development
|
0.0190744 |
0.0190744 |
1
|
10
|
|
GO:0003824
|
catalytic activity
|
0.0291395 |
0.0291395 |
5
|
4106
|
|
GO:0008065
|
establishment of blood-nerve barrier
|
0.0300000 |
0.0300000 |
1
|
3
|
|
GO:0048869
|
cellular developmental process
|
0.0312311 |
0.0312311 |
2
|
1276
|
|
GO:0030855
|
epithelial cell differentiation
|
0.0350466 |
0.0350466 |
1
|
24
|
|
GO:0003001
|
generation of a signal involved in cell-cell signaling
|
0.0377495 |
0.0377495 |
1
|
125
|
|
GO:0032940
|
secretion by cell
|
0.0433846 |
0.0433846 |
1
|
144
|
|
GO:0008544
|
epidermis development
|
0.0438533 |
0.0438533 |
1
|
61
|
|
GO:0035321
|
maintenance of imaginal disc-derived wing hair orientation
|
0.0476190 |
0.0476190 |
1
|
3
|
|
GO:0016458
|
gene silencing
|
0.0490740 |
0.0490740 |
1
|
163
|
|
GO:0009954
|
proximal/distal pattern formation
|
0.0494071 |
0.0494071 |
1
|
25
|
|
GO:0016020
|
membrane
|
0.0499001 |
0.0499001 |
4
|
2122
|
|
GO:0007059
|
chromosome segregation
|
0.0499732 |
0.0499732 |
1
|
166
|
|
GO:0001882
|
nucleoside binding
|
0.0525332 |
0.0525332 |
2
|
784
|
|
GO:0004091
|
carboxylesterase activity
|
0.0543337 |
0.0543337 |
2
|
116
|
|
GO:0016627
|
oxidoreductase activity, acting on the CH-CH group of donors
|
0.0579937 |
0.0579937 |
1
|
37
|
|
GO:0007413
|
axonal fasciculation
|
0.0627306 |
0.0627306 |
1
|
17
|
|
GO:0034329
|
cell junction assembly
|
0.0660870 |
0.0660870 |
1
|
38
|
|
GO:0043190
|
ATP-binding cassette (ABC) transporter complex
|
0.0665139 |
0.0665139 |
1
|
43
|
|
GO:0009913
|
epidermal cell differentiation
|
0.0726163 |
0.0726163 |
1
|
46
|
|
GO:0006457
|
protein folding
|
0.0757475 |
0.0757475 |
1
|
114
|
|
GO:0044425
|
membrane part
|
0.0794503 |
0.0794503 |
3
|
1398
|
|
GO:0065008
|
regulation of biological quality
|
0.0817769 |
0.0817769 |
2
|
729
|
|
GO:0051082
|
unfolded protein binding
|
0.0861504 |
0.0861504 |
1
|
76
|
|
GO:0010605
|
negative regulation of macromolecule metabolic process
|
0.0988340 |
0.0988340 |
1
|
356
|
|
GO:0016321
|
female meiosis chromosome segregation
|
0.103817 |
0.103817 |
1
|
68
|
|
GO:0000166
|
nucleotide binding
|
0.112563 |
0.112563 |
2
|
1178
|
|
GO:0007422
|
peripheral nervous system development
|
0.114481 |
0.114481 |
1
|
100
|
|
GO:0008038
|
neuron recognition
|
0.118644 |
0.118644 |
1
|
56
|
|
GO:0045446
|
endothelial cell differentiation
|
0.125000 |
0.125000 |
1
|
3
|
|
GO:0006629
|
lipid metabolic process
|
0.132436 |
0.132436 |
1
|
292
|
|
GO:0010608
|
posttranscriptional regulation of gene expression
|
0.133607 |
0.133607 |
1
|
130
|
|
GO:0009892
|
negative regulation of metabolic process
|
0.133617 |
0.133617 |
1
|
372
|
|
GO:0005811
|
lipid particle
|
0.134192 |
0.134192 |
1
|
250
|
|
GO:0007154
|
cell communication
|
0.135620 |
0.135620 |
1
|
461
|
|
GO:0016327
|
apicolateral plasma membrane
|
0.136729 |
0.136729 |
1
|
51
|
|
GO:0010629
|
negative regulation of gene expression
|
0.149146 |
0.149146 |
1
|
288
|
|
GO:0035152
|
regulation of tube architecture, open tracheal system
|
0.150424 |
0.150424 |
1
|
69
|
|
GO:0005739
|
mitochondrion
|
0.153911 |
0.153911 |
1
|
551
|
|
GO:0051234
|
establishment of localization
|
0.157272 |
0.157272 |
2
|
1549
|
|
GO:0016441
|
posttranscriptional gene silencing
|
0.160000 |
0.160000 |
1
|
44
|
|
GO:0016787
|
hydrolase activity
|
0.163662 |
0.163662 |
4
|
1972
|
|
GO:0023061
|
signal release
|
0.164474 |
0.164474 |
1
|
125
|
|
GO:0005681
|
spliceosomal complex
|
0.167598 |
0.167598 |
1
|
180
|
|
GO:0035317
|
imaginal disc-derived wing hair organization
|
0.168627 |
0.168627 |
1
|
43
|
|
GO:0005623
|
cell
|
0.171157 |
0.171157 |
5
|
6195
|
|
GO:0044464
|
cell part
|
0.171157 |
0.171157 |
5
|
6195
|
|
GO:0035316
|
non-sensory hair organization
|
0.173383 |
0.173383 |
1
|
44
|
|
GO:0016192
|
vesicle-mediated transport
|
0.176723 |
0.176723 |
1
|
430
|
|
GO:0040029
|
regulation of gene expression, epigenetic
|
0.181912 |
0.181912 |
1
|
177
|
|
GO:0006836
|
neurotransmitter transport
|
0.185604 |
0.185604 |
1
|
146
|
|
GO:0007267
|
cell-cell signaling
|
0.188801 |
0.188801 |
1
|
263
|
|
GO:0022402
|
cell cycle process
|
0.189739 |
0.189739 |
1
|
655
|
|
GO:0046903
|
secretion
|
0.192824 |
0.192824 |
1
|
152
|
|
GO:0019226
|
transmission of nerve impulse
|
0.194826 |
0.194826 |
1
|
241
|
|
GO:0040011
|
locomotion
|
0.208134 |
0.208134 |
1
|
484
|
|
GO:0048468
|
cell development
|
0.209123 |
0.209123 |
2
|
1042
|
|
GO:0048736
|
appendage development
|
0.209532 |
0.209532 |
1
|
286
|
|
GO:0006396
|
RNA processing
|
0.214286 |
0.214286 |
1
|
414
|
|
GO:0007049
|
cell cycle
|
0.217222 |
0.217222 |
1
|
756
|
|
GO:0051179
|
localization
|
0.219545 |
0.219545 |
2
|
1896
|
|
GO:0016820
|
hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances
|
0.221629 |
0.221629 |
1
|
139
|
|
GO:0005515
|
protein binding
|
0.223526 |
0.223526 |
2
|
1726
|
|
GO:0044430
|
cytoskeletal part
|
0.231674 |
0.231674 |
1
|
512
|
|
GO:0009605
|
response to external stimulus
|
0.242248 |
0.242248 |
1
|
375
|
|
GO:0051641
|
cellular localization
|
0.242560 |
0.242560 |
1
|
607
|
|
GO:0007399
|
nervous system development
|
0.249853 |
0.249853 |
2
|
848
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.259771 |
0.259771 |
2
|
2108
|
|
GO:0016788
|
hydrolase activity, acting on ester bonds
|
0.264183 |
0.264183 |
2
|
496
|
|
GO:0030054
|
cell junction
|
0.270777 |
0.270777 |
1
|
101
|
|
GO:0044428
|
nuclear part
|
0.271419 |
0.271419 |
1
|
830
|
|
GO:0060541
|
respiratory system development
|
0.272151 |
0.272151 |
1
|
249
|
|
GO:0035150
|
regulation of tube size
|
0.272189 |
0.272189 |
1
|
46
|
|
GO:0048569
|
post-embryonic organ development
|
0.276167 |
0.276167 |
1
|
343
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.283034 |
0.283034 |
1
|
1056
|
|
GO:0016071
|
mRNA metabolic process
|
0.284635 |
0.284635 |
1
|
339
|
|
GO:0006935
|
chemotaxis
|
0.287630 |
0.287630 |
1
|
193
|
|
GO:0048519
|
negative regulation of biological process
|
0.291182 |
0.291182 |
1
|
706
|
|
GO:0005737
|
cytoplasm
|
0.294170 |
0.294170 |
2
|
2378
|
|
GO:0031047
|
gene silencing by RNA
|
0.294479 |
0.294479 |
1
|
48
|
|
GO:0030529
|
ribonucleoprotein complex
|
0.298647 |
0.298647 |
1
|
510
|
|
GO:0001885
|
endothelial cell development
|
0.300000 |
0.300000 |
1
|
3
|
|
GO:0001505
|
regulation of neurotransmitter levels
|
0.304305 |
0.304305 |
1
|
129
|
|
GO:0023046
|
signaling process
|
0.310289 |
0.310289 |
1
|
760
|
|
GO:0045132
|
meiotic chromosome segregation
|
0.315287 |
0.315287 |
1
|
99
|
|
GO:0035107
|
appendage morphogenesis
|
0.334837 |
0.334837 |
1
|
283
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.338051 |
0.338051 |
2
|
1534
|
|
GO:0009791
|
post-embryonic development
|
0.343209 |
0.343209 |
1
|
500
|
|
GO:0065007
|
biological regulation
|
0.346129 |
0.346129 |
2
|
2548
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.346904 |
0.346904 |
1
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.347592 |
0.347592 |
1
|
1227
|
|
GO:0032989
|
cellular component morphogenesis
|
0.348979 |
0.348979 |
1
|
566
|
|
GO:0016817
|
hydrolase activity, acting on acid anhydrides
|
0.350404 |
0.350404 |
2
|
594
|
|
GO:0032991
|
macromolecular complex
|
0.354447 |
0.354447 |
2
|
2151
|
|
GO:0051321
|
meiotic cell cycle
|
0.357143 |
0.357143 |
1
|
270
|
|
GO:0003008
|
system process
|
0.360531 |
0.360531 |
1
|
664
|
|
GO:0032502
|
developmental process
|
0.364008 |
0.364008 |
2
|
2638
|
|
GO:0007389
|
pattern specification process
|
0.365750 |
0.365750 |
1
|
537
|
|
GO:0016887
|
ATPase activity
|
0.371288 |
0.371288 |
2
|
353
|
|
GO:0071844
|
cellular component assembly at cellular level
|
0.373193 |
0.373193 |
1
|
568
|
|
GO:0009792
|
embryo development ending in birth or egg hatching
|
0.374415 |
0.374415 |
1
|
240
|
|
GO:0007444
|
imaginal disc development
|
0.376006 |
0.376006 |
1
|
467
|
|
GO:0019222
|
regulation of metabolic process
|
0.376562 |
0.376562 |
1
|
1242
|
|
GO:0005215
|
transporter activity
|
0.381781 |
0.381781 |
1
|
852
|
|
GO:0042330
|
taxis
|
0.389803 |
0.389803 |
1
|
237
|
|
GO:0005875
|
microtubule associated complex
|
0.399123 |
0.399123 |
1
|
363
|
|
GO:0007143
|
female meiosis
|
0.401575 |
0.401575 |
1
|
102
|
|
GO:0005575
|
cellular_component
|
0.402358 |
0.402358 |
5
|
8817
|
|
GO:0042221
|
response to chemical stimulus
|
0.408269 |
0.408269 |
1
|
632
|
|
GO:0090066
|
regulation of anatomical structure size
|
0.410151 |
0.410151 |
1
|
169
|
|
GO:0016070
|
RNA metabolic process
|
0.413829 |
0.413829 |
1
|
1191
|
|
GO:0048563
|
post-embryonic organ morphogenesis
|
0.415167 |
0.415167 |
1
|
323
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.418447 |
0.418447 |
1
|
1562
|
|
GO:0009790
|
embryo development
|
0.427001 |
0.427001 |
1
|
641
|
|
GO:0003674
|
molecular_function
|
0.427511 |
0.427511 |
6
|
11064
|
|
GO:0009888
|
tissue development
|
0.431439 |
0.431439 |
1
|
557
|
|
GO:0048731
|
system development
|
0.432376 |
0.432376 |
2
|
1696
|
|
GO:0007126
|
meiosis
|
0.432709 |
0.432709 |
1
|
254
|
|
GO:0043492
|
ATPase activity, coupled to movement of substances
|
0.437107 |
0.437107 |
1
|
139
|
|
GO:0044267
|
cellular protein metabolic process
|
0.440058 |
0.440058 |
1
|
1505
|
|
GO:0007275
|
multicellular organismal development
|
0.442066 |
0.442066 |
2
|
2296
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.451701 |
0.451701 |
1
|
412
|
|
GO:0015399
|
primary active transmembrane transporter activity
|
0.452778 |
0.452778 |
1
|
163
|
|
GO:0051327
|
M phase of meiotic cell cycle
|
0.456014 |
0.456014 |
1
|
254
|
|
GO:0005856
|
cytoskeleton
|
0.457213 |
0.457213 |
1
|
561
|
|
GO:0071944
|
cell periphery
|
0.462926 |
0.462926 |
1
|
724
|
|
GO:0007552
|
metamorphosis
|
0.472755 |
0.472755 |
1
|
420
|
|
GO:0022607
|
cellular component assembly
|
0.475076 |
0.475076 |
1
|
577
|
|
GO:0070160
|
occluding junction
|
0.476190 |
0.476190 |
1
|
30
|
|
GO:0010468
|
regulation of gene expression
|
0.489930 |
0.489930 |
1
|
973
|
|
GO:0022804
|
active transmembrane transporter activity
|
0.495868 |
0.495868 |
1
|
360
|
|
GO:0032501
|
multicellular organismal process
|
0.496887 |
0.496887 |
2
|
3315
|
|
GO:0023052
|
signaling
|
0.497295 |
0.497295 |
1
|
1364
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.498346 |
0.498346 |
1
|
1959
|
|
GO:0044085
|
cellular component biogenesis
|
0.509834 |
0.509834 |
1
|
632
|
|
GO:0051649
|
establishment of localization in cell
|
0.515645 |
0.515645 |
1
|
500
|
|
GO:0043234
|
protein complex
|
0.528587 |
0.528587 |
2
|
1564
|
|
GO:0060429
|
epithelium development
|
0.529623 |
0.529623 |
1
|
295
|
|
GO:0010467
|
gene expression
|
0.531494 |
0.531494 |
1
|
1907
|
|
GO:0030018
|
Z disc
|
0.538462 |
0.538462 |
1
|
14
|
|
GO:0031674
|
I band
|
0.538462 |
0.538462 |
1
|
14
|
|
GO:0005918
|
septate junction
|
0.543478 |
0.543478 |
1
|
25
|
|
GO:0050896
|
response to stimulus
|
0.545344 |
0.545344 |
1
|
1548
|
|
GO:0044459
|
plasma membrane part
|
0.555500 |
0.555500 |
1
|
373
|
|
GO:0032553
|
ribonucleotide binding
|
0.556624 |
0.556624 |
2
|
879
|
|
GO:0016491
|
oxidoreductase activity
|
0.570359 |
0.570359 |
1
|
638
|
|
GO:0032559
|
adenyl ribonucleotide binding
|
0.573921 |
0.573921 |
2
|
713
|
|
GO:0007560
|
imaginal disc morphogenesis
|
0.575758 |
0.575758 |
1
|
323
|
|
GO:0009887
|
organ morphogenesis
|
0.608733 |
0.608733 |
1
|
684
|
|
GO:0000904
|
cell morphogenesis involved in differentiation
|
0.611753 |
0.611753 |
1
|
408
|
|
GO:0005911
|
cell-cell junction
|
0.623762 |
0.623762 |
1
|
63
|
|
GO:0035151
|
regulation of tube size, open tracheal system
|
0.628571 |
0.628571 |
1
|
44
|
|
GO:0035120
|
post-embryonic appendage morphogenesis
|
0.638095 |
0.638095 |
1
|
268
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.639129 |
0.639129 |
1
|
2878
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.643595 |
0.643595 |
1
|
2093
|
|
GO:0017076
|
purine nucleotide binding
|
0.644765 |
0.644765 |
2
|
946
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.658716 |
0.658716 |
1
|
1789
|
|
GO:0007472
|
wing disc morphogenesis
|
0.665796 |
0.665796 |
1
|
255
|
|
GO:0008380
|
RNA splicing
|
0.669082 |
0.669082 |
1
|
277
|
|
GO:0044444
|
cytoplasmic part
|
0.669378 |
0.669378 |
1
|
1863
|
|
GO:0030554
|
adenyl nucleotide binding
|
0.670996 |
0.670996 |
2
|
775
|
|
GO:0044238
|
primary metabolic process
|
0.672346 |
0.672346 |
2
|
4259
|
|
GO:0005488
|
binding
|
0.674581 |
0.674581 |
3
|
5641
|
|
GO:0007409
|
axonogenesis
|
0.679389 |
0.679389 |
1
|
267
|
|
GO:0048666
|
neuron development
|
0.679533 |
0.679533 |
1
|
471
|
|
GO:0035220
|
wing disc development
|
0.680942 |
0.680942 |
1
|
318
|
|
GO:0071013
|
catalytic step 2 spliceosome
|
0.683333 |
0.683333 |
1
|
123
|
|
GO:0007411
|
axon guidance
|
0.683824 |
0.683824 |
1
|
186
|
|
GO:0043296
|
apical junction complex
|
0.686567 |
0.686567 |
1
|
46
|
|
GO:0030182
|
neuron differentiation
|
0.689161 |
0.689161 |
1
|
548
|
|
GO:0019991
|
septate junction assembly
|
0.694444 |
0.694444 |
1
|
25
|
|
GO:0050789
|
regulation of biological process
|
0.695412 |
0.695412 |
1
|
2246
|
|
GO:0022008
|
neurogenesis
|
0.700310 |
0.700310 |
1
|
632
|
|
GO:0007269
|
neurotransmitter secretion
|
0.701149 |
0.701149 |
1
|
122
|
|
GO:0016246
|
RNA interference
|
0.704545 |
0.704545 |
1
|
31
|
|
GO:0019538
|
protein metabolic process
|
0.724907 |
0.724907 |
1
|
2053
|
|
GO:0006397
|
mRNA processing
|
0.734411 |
0.734411 |
1
|
318
|
|
GO:0008150
|
biological_process
|
0.735561 |
0.735561 |
5
|
10616
|
|
GO:0048856
|
anatomical structure development
|
0.737157 |
0.737157 |
2
|
2265
|
|
GO:0044446
|
intracellular organelle part
|
0.743053 |
0.743053 |
1
|
2162
|
|
GO:0032990
|
cell part morphogenesis
|
0.745583 |
0.745583 |
1
|
422
|
|
GO:0005886
|
plasma membrane
|
0.749057 |
0.749057 |
1
|
641
|
|
GO:0044422
|
organelle part
|
0.757673 |
0.757673 |
1
|
2176
|
|
GO:0007043
|
cell-cell junction assembly
|
0.765957 |
0.765957 |
1
|
36
|
|
GO:0048667
|
cell morphogenesis involved in neuron differentiation
|
0.795501 |
0.795501 |
1
|
389
|
|
GO:0048513
|
organ development
|
0.796117 |
0.796117 |
1
|
1242
|
|
GO:0015630
|
microtubule cytoskeleton
|
0.798574 |
0.798574 |
1
|
448
|
|
GO:0008152
|
metabolic process
|
0.798847 |
0.798847 |
2
|
5194
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0031224
|
intrinsic to membrane
|
0.815297 |
0.815297 |
2
|
1014
|
|
GO:0071011
|
precatalytic spliceosome
|
0.822222 |
0.822222 |
1
|
148
|
|
GO:0000902
|
cell morphogenesis
|
0.825088 |
0.825088 |
1
|
467
|
|
GO:0000398
|
nuclear mRNA splicing, via spliceosome
|
0.836478 |
0.836478 |
1
|
266
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.842767 |
0.842767 |
1
|
402
|
|
GO:0022857
|
transmembrane transporter activity
|
0.852113 |
0.852113 |
1
|
726
|
|
GO:0042626
|
ATPase activity, coupled to transmembrane movement of substances
|
0.852761 |
0.852761 |
1
|
139
|
|
GO:0030016
|
myofibril
|
0.857143 |
0.857143 |
1
|
24
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.863297 |
0.863297 |
2
|
879
|
|
GO:0007268
|
synaptic transmission
|
0.866667 |
0.866667 |
1
|
234
|
|
GO:0044237
|
cellular metabolic process
|
0.868168 |
0.868168 |
1
|
3814
|
|
GO:0030017
|
sarcomere
|
0.884615 |
0.884615 |
1
|
23
|
|
GO:0022403
|
cell cycle phase
|
0.896183 |
0.896183 |
1
|
587
|
|
GO:0007476
|
imaginal disc-derived wing morphogenesis
|
0.903915 |
0.903915 |
1
|
254
|
|
GO:0043170
|
macromolecule metabolic process
|
0.904435 |
0.904435 |
1
|
3588
|
|
GO:0031175
|
neuron projection development
|
0.908124 |
0.908124 |
1
|
392
|
|
GO:0048812
|
neuron projection morphogenesis
|
0.910798 |
0.910798 |
1
|
388
|
|
GO:0035194
|
posttranscriptional gene silencing by RNA
|
0.916667 |
0.916667 |
1
|
44
|
|
GO:0043226
|
organelle
|
0.923045 |
0.923045 |
1
|
3537
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.923977 |
0.923977 |
1
|
1517
|
|
GO:0009987
|
cellular process
|
0.926071 |
0.926071 |
2
|
6560
|
|
GO:0006810
|
transport
|
0.933966 |
0.933966 |
2
|
1497
|
|
GO:0003002
|
regionalization
|
0.942272 |
0.942272 |
1
|
506
|
|
GO:0030154
|
cell differentiation
|
0.942825 |
0.942825 |
2
|
1239
|
|
GO:0000279
|
M phase
|
0.945486 |
0.945486 |
1
|
555
|
|
GO:0016043
|
cellular component organization
|
0.947563 |
0.947563 |
2
|
2052
|
|
GO:0050877
|
neurological system process
|
0.953313 |
0.953313 |
1
|
633
|
|
GO:0002165
|
instar larval or pupal development
|
0.954000 |
0.954000 |
1
|
477
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0035315
|
hair cell differentiation
|
0.956522 |
0.956522 |
1
|
44
|
|
GO:0045216
|
cell-cell junction organization
|
0.957447 |
0.957447 |
1
|
45
|
|
GO:0001700
|
embryonic development via the syncytial blastoderm
|
0.958333 |
0.958333 |
1
|
230
|
|
GO:0043229
|
intracellular organelle
|
0.961659 |
0.961659 |
1
|
3530
|
|
GO:0048858
|
cell projection morphogenesis
|
0.963004 |
0.963004 |
1
|
422
|
|
GO:0000375
|
RNA splicing, via transesterification reactions
|
0.963899 |
0.963899 |
1
|
267
|
|
GO:0017111
|
nucleoside-triphosphatase activity
|
0.966304 |
0.966304 |
2
|
579
|
|
GO:0016021
|
integral to membrane
|
0.968675 |
0.968675 |
2
|
998
|
|
GO:0044424
|
intracellular part
|
0.972076 |
0.972076 |
2
|
4384
|
|
GO:0043297
|
apical junction assembly
|
0.972222 |
0.972222 |
1
|
35
|
|
GO:0035114
|
imaginal disc-derived appendage morphogenesis
|
0.975524 |
0.975524 |
1
|
279
|
|
GO:0016462
|
pyrophosphatase activity
|
0.983222 |
0.983222 |
2
|
589
|
|
GO:0001883
|
purine nucleoside binding
|
0.984743 |
0.984743 |
2
|
778
|
|
GO:0048737
|
imaginal disc-derived appendage development
|
0.986014 |
0.986014 |
1
|
282
|
|
GO:0005622
|
intracellular
|
0.987484 |
0.987484 |
2
|
4734
|
|
GO:0042623
|
ATPase activity, coupled
|
0.990423 |
0.990423 |
1
|
318
|
|
GO:0007424
|
open tracheal system development
|
0.995984 |
0.995984 |
1
|
248
|
|
GO:0000377
|
RNA splicing, via transesterification reactions with bulged adenosine as nucleophile
|
0.996255 |
0.996255 |
1
|
266
|
|
GO:0005524
|
ATP binding
|
0.997195 |
0.997195 |
2
|
712
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
6
|
12747
|
|
GO:0016818
|
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
|
1.00000 |
1.00000 |
2
|
594
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|
|
GO:0015405
|
P-P-bond-hydrolysis-driven transmembrane transporter activity
|
1.00000 |
1.00000 |
1
|
163
|