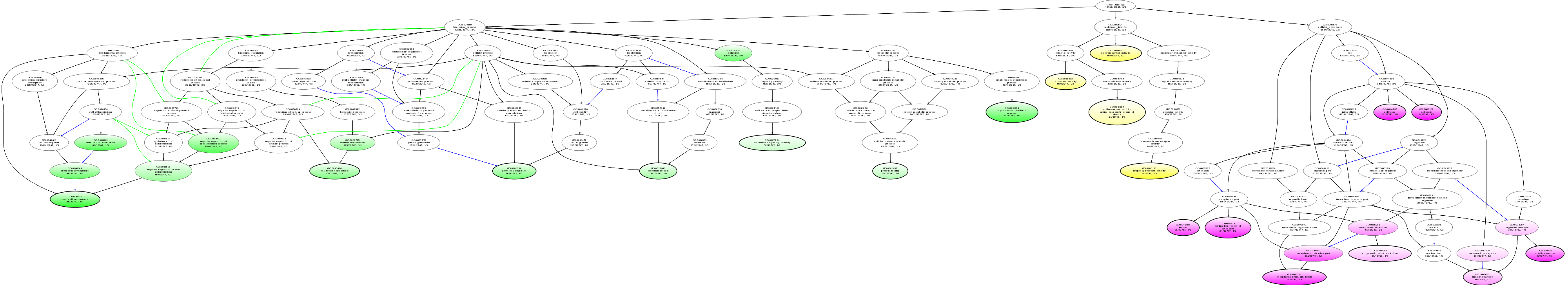
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0060187
|
cell pole
|
0.000968367 |
0.000968367 |
1
|
2
|
|
GO:0044297
|
cell body
|
0.0110985 |
0.0110985 |
1
|
23
|
|
GO:0005788
|
endoplasmic reticulum lumen
|
0.0123457 |
0.0123457 |
1
|
9
|
|
GO:0048471
|
perinuclear region of cytoplasm
|
0.0128824 |
0.0128824 |
1
|
24
|
|
GO:0070732
|
spindle envelope
|
0.0164474 |
0.0164474 |
1
|
5
|
|
GO:0019827
|
stem cell maintenance
|
0.0174375 |
0.0174375 |
1
|
46
|
|
GO:0008158
|
hedgehog receptor activity
|
0.0177215 |
0.0177215 |
1
|
7
|
|
GO:0045169
|
fusome
|
0.0193237 |
0.0193237 |
1
|
36
|
|
GO:0018904
|
organic ether metabolic process
|
0.0328515 |
0.0328515 |
1
|
25
|
|
GO:0044432
|
endoplasmic reticulum part
|
0.0339315 |
0.0339315 |
1
|
104
|
|
GO:0048863
|
stem cell differentiation
|
0.0371267 |
0.0371267 |
1
|
46
|
|
GO:0051093
|
negative regulation of developmental process
|
0.0423208 |
0.0423208 |
1
|
124
|
|
GO:0009055
|
electron carrier activity
|
0.0438166 |
0.0438166 |
1
|
164
|
|
GO:0048864
|
stem cell development
|
0.0441459 |
0.0441459 |
1
|
46
|
|
GO:0023052
|
signaling
|
0.0452597 |
0.0452597 |
2
|
1364
|
|
GO:0008354
|
germ cell migration
|
0.0472279 |
0.0472279 |
1
|
46
|
|
GO:0016853
|
isomerase activity
|
0.0476465 |
0.0476465 |
1
|
99
|
|
GO:0045454
|
cell redox homeostasis
|
0.0520496 |
0.0520496 |
1
|
55
|
|
GO:0005783
|
endoplasmic reticulum
|
0.0555866 |
0.0555866 |
1
|
199
|
|
GO:0019725
|
cellular homeostasis
|
0.0560655 |
0.0560655 |
1
|
125
|
|
GO:0005635
|
nuclear envelope
|
0.0591716 |
0.0591716 |
1
|
70
|
|
GO:0045596
|
negative regulation of cell differentiation
|
0.0604154 |
0.0604154 |
1
|
96
|
|
GO:0032940
|
secretion by cell
|
0.0643707 |
0.0643707 |
1
|
144
|
|
GO:0016667
|
oxidoreductase activity, acting on a sulfur group of donors
|
0.0689655 |
0.0689655 |
1
|
44
|
|
GO:0005791
|
rough endoplasmic reticulum
|
0.0753769 |
0.0753769 |
1
|
15
|
|
GO:0006457
|
protein folding
|
0.0757475 |
0.0757475 |
1
|
114
|
|
GO:0031967
|
organelle envelope
|
0.0883978 |
0.0883978 |
1
|
304
|
|
GO:0012505
|
endomembrane system
|
0.0896856 |
0.0896856 |
1
|
191
|
|
GO:0007224
|
smoothened signaling pathway
|
0.0929487 |
0.0929487 |
1
|
58
|
|
GO:0046903
|
secretion
|
0.101536 |
0.101536 |
1
|
152
|
|
GO:0050789
|
regulation of biological process
|
0.115315 |
0.115315 |
2
|
2246
|
|
GO:0006928
|
cellular component movement
|
0.118060 |
0.118060 |
1
|
269
|
|
GO:0040011
|
locomotion
|
0.130645 |
0.130645 |
1
|
484
|
|
GO:0005811
|
lipid particle
|
0.134192 |
0.134192 |
1
|
250
|
|
GO:0051674
|
localization of cell
|
0.138713 |
0.138713 |
1
|
263
|
|
GO:0060089
|
molecular transducer activity
|
0.144056 |
0.144056 |
1
|
559
|
|
GO:0031975
|
envelope
|
0.144503 |
0.144503 |
1
|
314
|
|
GO:0065007
|
biological regulation
|
0.145142 |
0.145142 |
2
|
2548
|
|
GO:0044281
|
small molecule metabolic process
|
0.146515 |
0.146515 |
1
|
761
|
|
GO:0050793
|
regulation of developmental process
|
0.161873 |
0.161873 |
1
|
321
|
|
GO:0006887
|
exocytosis
|
0.167038 |
0.167038 |
1
|
75
|
|
GO:0045595
|
regulation of cell differentiation
|
0.167859 |
0.167859 |
1
|
231
|
|
GO:0016192
|
vesicle-mediated transport
|
0.176723 |
0.176723 |
1
|
430
|
|
GO:0019001
|
guanyl nucleotide binding
|
0.182875 |
0.182875 |
1
|
173
|
|
GO:0048519
|
negative regulation of biological process
|
0.186553 |
0.186553 |
1
|
706
|
|
GO:0032561
|
guanyl ribonucleotide binding
|
0.194098 |
0.194098 |
1
|
171
|
|
GO:0023046
|
signaling process
|
0.199779 |
0.199779 |
1
|
760
|
|
GO:0031974
|
membrane-enclosed lumen
|
0.204405 |
0.204405 |
1
|
647
|
|
GO:0000166
|
nucleotide binding
|
0.208828 |
0.208828 |
1
|
1178
|
|
GO:0050794
|
regulation of cellular process
|
0.224039 |
0.224039 |
2
|
2034
|
|
GO:0009987
|
cellular process
|
0.235914 |
0.235914 |
3
|
6560
|
|
GO:0003924
|
GTPase activity
|
0.241796 |
0.241796 |
1
|
140
|
|
GO:0051641
|
cellular localization
|
0.242560 |
0.242560 |
1
|
607
|
|
GO:0048610
|
cellular process involved in reproduction
|
0.248418 |
0.248418 |
1
|
613
|
|
GO:0042592
|
homeostatic process
|
0.256516 |
0.256516 |
1
|
187
|
|
GO:0000003
|
reproduction
|
0.261920 |
0.261920 |
1
|
1022
|
|
GO:0022414
|
reproductive process
|
0.261920 |
0.261920 |
1
|
1022
|
|
GO:0048523
|
negative regulation of cellular process
|
0.262869 |
0.262869 |
1
|
635
|
|
GO:0044428
|
nuclear part
|
0.271419 |
0.271419 |
1
|
830
|
|
GO:0016020
|
membrane
|
0.271576 |
0.271576 |
2
|
2122
|
|
GO:0048609
|
multicellular organismal reproductive process
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0032504
|
multicellular organism reproduction
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0016491
|
oxidoreductase activity
|
0.286653 |
0.286653 |
1
|
638
|
|
GO:0043233
|
organelle lumen
|
0.291360 |
0.291360 |
1
|
634
|
|
GO:0016860
|
intramolecular oxidoreductase activity
|
0.292929 |
0.292929 |
1
|
29
|
|
GO:0070013
|
intracellular organelle lumen
|
0.293247 |
0.293247 |
1
|
634
|
|
GO:0016817
|
hydrolase activity, acting on acid anhydrides
|
0.301217 |
0.301217 |
1
|
594
|
|
GO:0051649
|
establishment of localization in cell
|
0.303951 |
0.303951 |
1
|
500
|
|
GO:0003824
|
catalytic activity
|
0.310936 |
0.310936 |
2
|
4106
|
|
GO:0005575
|
cellular_component
|
0.330897 |
0.330897 |
3
|
8817
|
|
GO:0005623
|
cell
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0044464
|
cell part
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0051234
|
establishment of localization
|
0.377001 |
0.377001 |
1
|
1549
|
|
GO:0016862
|
intramolecular oxidoreductase activity, interconverting keto- and enol-groups
|
0.379310 |
0.379310 |
1
|
11
|
|
GO:0016864
|
intramolecular oxidoreductase activity, transposing S-S bonds
|
0.379310 |
0.379310 |
1
|
11
|
|
GO:0044444
|
cytoplasmic part
|
0.424954 |
0.424954 |
1
|
1863
|
|
GO:0007280
|
pole cell migration
|
0.434783 |
0.434783 |
1
|
20
|
|
GO:0044267
|
cellular protein metabolic process
|
0.440058 |
0.440058 |
1
|
1505
|
|
GO:0048869
|
cellular developmental process
|
0.442161 |
0.442161 |
1
|
1276
|
|
GO:0007165
|
signal transduction
|
0.443656 |
0.443656 |
1
|
548
|
|
GO:0051179
|
localization
|
0.445834 |
0.445834 |
1
|
1896
|
|
GO:0048468
|
cell development
|
0.457419 |
0.457419 |
1
|
1042
|
|
GO:0019538
|
protein metabolic process
|
0.475452 |
0.475452 |
1
|
2053
|
|
GO:0065008
|
regulation of biological quality
|
0.490437 |
0.490437 |
1
|
729
|
|
GO:0023033
|
signaling pathway
|
0.491079 |
0.491079 |
2
|
956
|
|
GO:0044446
|
intracellular organelle part
|
0.493044 |
0.493044 |
1
|
2162
|
|
GO:0035556
|
intracellular signal transduction
|
0.499208 |
0.499208 |
1
|
315
|
|
GO:0048870
|
cell motility
|
0.512097 |
0.512097 |
1
|
254
|
|
GO:0044425
|
membrane part
|
0.535780 |
0.535780 |
1
|
1398
|
|
GO:0005737
|
cytoplasm
|
0.542427 |
0.542427 |
1
|
2378
|
|
GO:0044422
|
organelle part
|
0.572743 |
0.572743 |
1
|
2176
|
|
GO:0032502
|
developmental process
|
0.575616 |
0.575616 |
1
|
2638
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0007264
|
small GTPase mediated signal transduction
|
0.587302 |
0.587302 |
1
|
185
|
|
GO:0015036
|
disulfide oxidoreductase activity
|
0.636364 |
0.636364 |
1
|
28
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.639129 |
0.639129 |
1
|
2878
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0032501
|
multicellular organismal process
|
0.674757 |
0.674757 |
1
|
3315
|
|
GO:0023034
|
intracellular signaling pathway
|
0.676453 |
0.676453 |
1
|
412
|
|
GO:0043170
|
macromolecule metabolic process
|
0.690797 |
0.690797 |
1
|
3588
|
|
GO:0031224
|
intrinsic to membrane
|
0.725322 |
0.725322 |
1
|
1014
|
|
GO:0016787
|
hydrolase activity
|
0.729944 |
0.729944 |
1
|
1972
|
|
GO:0032553
|
ribonucleotide binding
|
0.746180 |
0.746180 |
1
|
879
|
|
GO:0015035
|
protein disulfide oxidoreductase activity
|
0.750000 |
0.750000 |
1
|
21
|
|
GO:0030718
|
germ-line stem cell maintenance
|
0.760870 |
0.760870 |
1
|
35
|
|
GO:0043226
|
organelle
|
0.785296 |
0.785296 |
1
|
3537
|
|
GO:0017076
|
purine nucleotide binding
|
0.803056 |
0.803056 |
1
|
946
|
|
GO:0043229
|
intracellular organelle
|
0.804100 |
0.804100 |
1
|
3530
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0044238
|
primary metabolic process
|
0.819985 |
0.819985 |
1
|
4259
|
|
GO:0004872
|
receptor activity
|
0.821109 |
0.821109 |
1
|
459
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0019953
|
sexual reproduction
|
0.859100 |
0.859100 |
1
|
878
|
|
GO:0005622
|
intracellular
|
0.859425 |
0.859425 |
2
|
4734
|
|
GO:0004888
|
transmembrane receptor activity
|
0.860566 |
0.860566 |
1
|
395
|
|
GO:0008152
|
metabolic process
|
0.866808 |
0.866808 |
1
|
5194
|
|
GO:0044237
|
cellular metabolic process
|
0.868168 |
0.868168 |
1
|
3814
|
|
GO:0007166
|
cell surface receptor linked signaling pathway
|
0.879634 |
0.879634 |
1
|
624
|
|
GO:0005488
|
binding
|
0.882277 |
0.882277 |
1
|
5641
|
|
GO:0007276
|
gamete generation
|
0.883697 |
0.883697 |
1
|
851
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.929175 |
0.929175 |
1
|
879
|
|
GO:0043025
|
neuronal cell body
|
0.956522 |
0.956522 |
1
|
22
|
|
GO:0016477
|
cell migration
|
0.964567 |
0.964567 |
1
|
245
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0044424
|
intracellular part
|
0.975047 |
0.975047 |
1
|
4384
|
|
GO:0017111
|
nucleoside-triphosphatase activity
|
0.983022 |
0.983022 |
1
|
579
|
|
GO:0016021
|
integral to membrane
|
0.984221 |
0.984221 |
1
|
998
|
|
GO:0016462
|
pyrophosphatase activity
|
0.991582 |
0.991582 |
1
|
589
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0016818
|
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
|
1.00000 |
1.00000 |
1
|
594
|
|
GO:0003756
|
protein disulfide isomerase activity
|
1.00000 |
1.00000 |
1
|
11
|
|
GO:0004871
|
signal transducer activity
|
1.00000 |
1.00000 |
1
|
559
|
|
GO:0005525
|
GTP binding
|
1.00000 |
1.00000 |
1
|
171
|
|
GO:0006662
|
glycerol ether metabolic process
|
1.00000 |
1.00000 |
1
|
25
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|