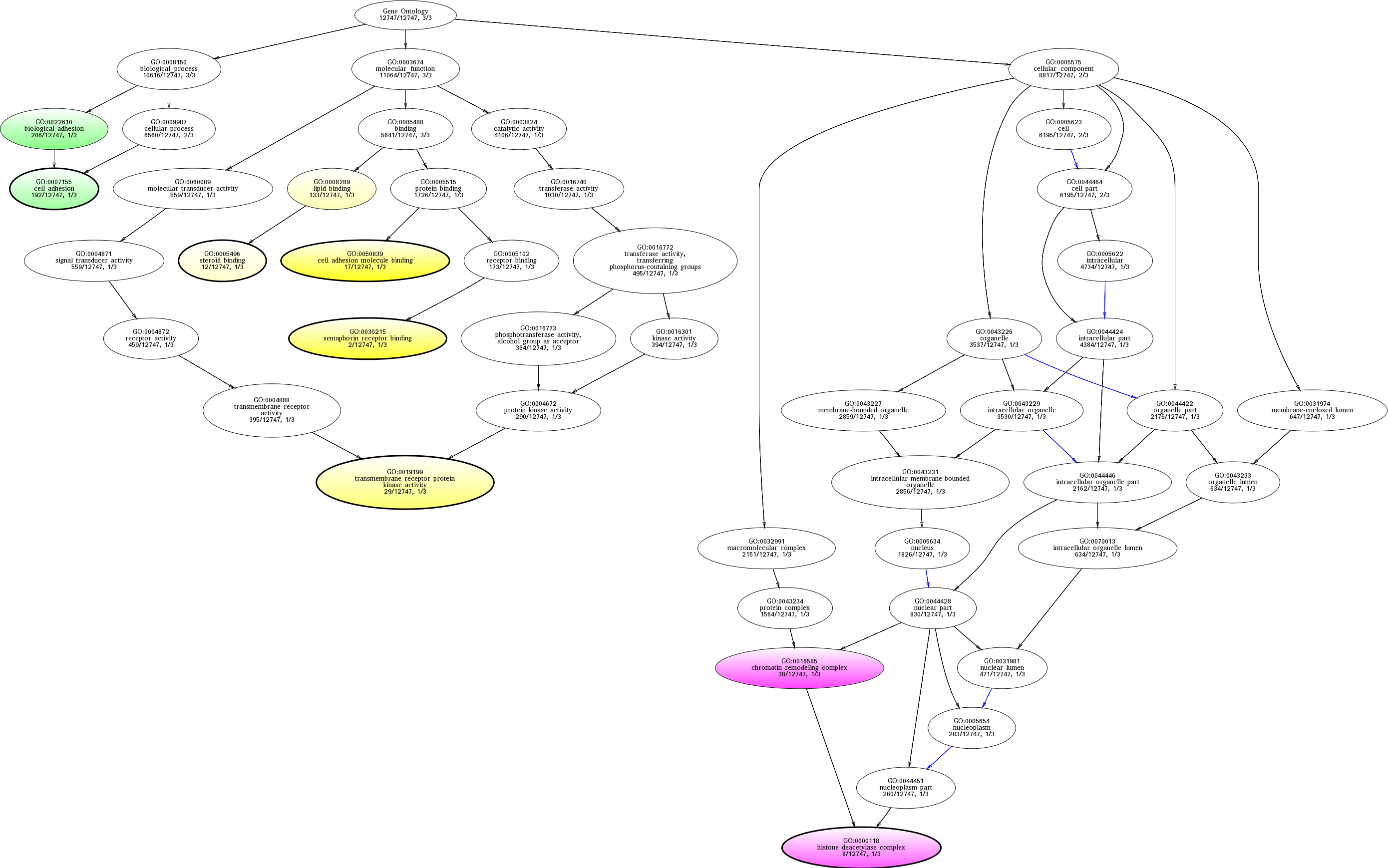
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0050839
|
cell adhesion molecule binding
|
0.00984936 |
0.00984936 |
1
|
17
|
|
GO:0030215
|
semaphorin receptor binding
|
0.0115607 |
0.0115607 |
1
|
2
|
|
GO:0016585
|
chromatin remodeling complex
|
0.0192016 |
0.0192016 |
1
|
38
|
|
GO:0000118
|
histone deacetylase complex
|
0.0277778 |
0.0277778 |
1
|
8
|
|
GO:0019199
|
transmembrane receptor protein kinase activity
|
0.0444104 |
0.0444104 |
1
|
29
|
|
GO:0022610
|
biological adhesion
|
0.0570970 |
0.0570970 |
1
|
206
|
|
GO:0007155
|
cell adhesion
|
0.0575719 |
0.0575719 |
1
|
192
|
|
GO:0008289
|
lipid binding
|
0.0690895 |
0.0690895 |
1
|
133
|
|
GO:0005496
|
steroid binding
|
0.0902256 |
0.0902256 |
1
|
12
|
|
GO:0005102
|
receptor binding
|
0.100232 |
0.100232 |
1
|
173
|
|
GO:0016044
|
cellular membrane organization
|
0.102136 |
0.102136 |
1
|
344
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.102602 |
0.102602 |
2
|
2108
|
|
GO:0008202
|
steroid metabolic process
|
0.102740 |
0.102740 |
1
|
30
|
|
GO:0001071
|
nucleic acid binding transcription factor activity
|
0.103335 |
0.103335 |
1
|
395
|
|
GO:0008152
|
metabolic process
|
0.117083 |
0.117083 |
3
|
5194
|
|
GO:0016192
|
vesicle-mediated transport
|
0.121581 |
0.121581 |
1
|
430
|
|
GO:0040011
|
locomotion
|
0.130645 |
0.130645 |
1
|
484
|
|
GO:0005488
|
binding
|
0.132501 |
0.132501 |
3
|
5641
|
|
GO:0031974
|
membrane-enclosed lumen
|
0.141385 |
0.141385 |
1
|
647
|
|
GO:0030030
|
cell projection organization
|
0.142264 |
0.142264 |
1
|
485
|
|
GO:0060089
|
molecular transducer activity
|
0.144056 |
0.144056 |
1
|
559
|
|
GO:0004713
|
protein tyrosine kinase activity
|
0.186207 |
0.186207 |
1
|
54
|
|
GO:0006629
|
lipid metabolic process
|
0.191945 |
0.191945 |
1
|
292
|
|
GO:0071944
|
cell periphery
|
0.220095 |
0.220095 |
1
|
724
|
|
GO:0009605
|
response to external stimulus
|
0.242248 |
0.242248 |
1
|
375
|
|
GO:0016740
|
transferase activity
|
0.250852 |
0.250852 |
1
|
1030
|
|
GO:0006793
|
phosphorus metabolic process
|
0.270339 |
0.270339 |
1
|
556
|
|
GO:0044428
|
nuclear part
|
0.271419 |
0.271419 |
1
|
830
|
|
GO:0006935
|
chemotaxis
|
0.287630 |
0.287630 |
1
|
193
|
|
GO:0032774
|
RNA biosynthetic process
|
0.290017 |
0.290017 |
1
|
703
|
|
GO:0043233
|
organelle lumen
|
0.291360 |
0.291360 |
1
|
634
|
|
GO:0005886
|
plasma membrane
|
0.292027 |
0.292027 |
1
|
641
|
|
GO:0070013
|
intracellular organelle lumen
|
0.293247 |
0.293247 |
1
|
634
|
|
GO:0043565
|
sequence-specific DNA binding
|
0.300971 |
0.300971 |
1
|
248
|
|
GO:0061024
|
membrane organization
|
0.308058 |
0.308058 |
1
|
345
|
|
GO:0044451
|
nucleoplasm part
|
0.313253 |
0.313253 |
1
|
260
|
|
GO:0048869
|
cellular developmental process
|
0.322329 |
0.322329 |
1
|
1276
|
|
GO:0006468
|
protein phosphorylation
|
0.335758 |
0.335758 |
1
|
277
|
|
GO:0005654
|
nucleoplasm
|
0.340964 |
0.340964 |
1
|
283
|
|
GO:0032989
|
cellular component morphogenesis
|
0.348979 |
0.348979 |
1
|
566
|
|
GO:0001882
|
nucleoside binding
|
0.361738 |
0.361738 |
1
|
784
|
|
GO:0043412
|
macromolecule modification
|
0.362896 |
0.362896 |
1
|
724
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.366791 |
0.366791 |
1
|
888
|
|
GO:0000904
|
cell morphogenesis involved in differentiation
|
0.376731 |
0.376731 |
1
|
408
|
|
GO:0050896
|
response to stimulus
|
0.376795 |
0.376795 |
1
|
1548
|
|
GO:0051234
|
establishment of localization
|
0.377001 |
0.377001 |
1
|
1549
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.381695 |
0.381695 |
1
|
905
|
|
GO:0006350
|
transcription
|
0.387810 |
0.387810 |
1
|
859
|
|
GO:0042330
|
taxis
|
0.389803 |
0.389803 |
1
|
237
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.396252 |
0.396252 |
1
|
888
|
|
GO:0008142
|
oxysterol binding
|
0.400000 |
0.400000 |
1
|
4
|
|
GO:0044425
|
membrane part
|
0.400435 |
0.400435 |
1
|
1398
|
|
GO:0004714
|
transmembrane receptor protein tyrosine kinase activity
|
0.406780 |
0.406780 |
1
|
24
|
|
GO:0042221
|
response to chemical stimulus
|
0.408269 |
0.408269 |
1
|
632
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.408435 |
0.408435 |
2
|
2878
|
|
GO:0032991
|
macromolecular complex
|
0.428425 |
0.428425 |
1
|
2151
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.429576 |
0.429576 |
1
|
1110
|
|
GO:0044422
|
organelle part
|
0.432705 |
0.432705 |
1
|
2176
|
|
GO:0048666
|
neuron development
|
0.433702 |
0.433702 |
1
|
471
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.440308 |
0.440308 |
1
|
686
|
|
GO:0030182
|
neuron differentiation
|
0.442292 |
0.442292 |
1
|
548
|
|
GO:0006464
|
protein modification process
|
0.443005 |
0.443005 |
1
|
684
|
|
GO:0003677
|
DNA binding
|
0.444205 |
0.444205 |
1
|
824
|
|
GO:0051179
|
localization
|
0.445834 |
0.445834 |
1
|
1896
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.451726 |
0.451726 |
1
|
903
|
|
GO:0022008
|
neurogenesis
|
0.452398 |
0.452398 |
1
|
632
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.456989 |
0.456989 |
1
|
850
|
|
GO:0048468
|
cell development
|
0.457419 |
0.457419 |
1
|
1042
|
|
GO:0031981
|
nuclear lumen
|
0.475277 |
0.475277 |
1
|
471
|
|
GO:0016772
|
transferase activity, transferring phosphorus-containing groups
|
0.480583 |
0.480583 |
1
|
495
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.486014 |
0.486014 |
1
|
1056
|
|
GO:0044237
|
cellular metabolic process
|
0.486483 |
0.486483 |
2
|
3814
|
|
GO:0010468
|
regulation of gene expression
|
0.489930 |
0.489930 |
1
|
973
|
|
GO:0044446
|
intracellular organelle part
|
0.493044 |
0.493044 |
1
|
2162
|
|
GO:0005623
|
cell
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0007399
|
nervous system development
|
0.500000 |
0.500000 |
1
|
848
|
|
GO:0000166
|
nucleotide binding
|
0.504833 |
0.504833 |
1
|
1178
|
|
GO:0019222
|
regulation of metabolic process
|
0.507779 |
0.507779 |
1
|
1242
|
|
GO:0050789
|
regulation of biological process
|
0.509927 |
0.509927 |
1
|
2246
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.513595 |
0.513595 |
1
|
850
|
|
GO:0050794
|
regulation of cellular process
|
0.518911 |
0.518911 |
1
|
2034
|
|
GO:0044238
|
primary metabolic process
|
0.551267 |
0.551267 |
3
|
4259
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.556274 |
0.556274 |
1
|
1039
|
|
GO:0065007
|
biological regulation
|
0.561089 |
0.561089 |
1
|
2548
|
|
GO:0016020
|
membrane
|
0.567775 |
0.567775 |
1
|
2122
|
|
GO:0032502
|
developmental process
|
0.575616 |
0.575616 |
1
|
2638
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0043167
|
ion binding
|
0.603911 |
0.603911 |
1
|
1498
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0043226
|
organelle
|
0.641414 |
0.641414 |
1
|
3537
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.646807 |
0.646807 |
1
|
1623
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0006897
|
endocytosis
|
0.655814 |
0.655814 |
1
|
282
|
|
GO:0016070
|
RNA metabolic process
|
0.656488 |
0.656488 |
1
|
1191
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0005515
|
protein binding
|
0.665786 |
0.665786 |
1
|
1726
|
|
GO:0009987
|
cellular process
|
0.673638 |
0.673638 |
2
|
6560
|
|
GO:0032501
|
multicellular organismal process
|
0.674757 |
0.674757 |
1
|
3315
|
|
GO:0007409
|
axonogenesis
|
0.679389 |
0.679389 |
1
|
267
|
|
GO:0006911
|
phagocytosis, engulfment
|
0.680851 |
0.680851 |
1
|
192
|
|
GO:0007411
|
axon guidance
|
0.683824 |
0.683824 |
1
|
186
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.684327 |
0.684327 |
1
|
620
|
|
GO:0044267
|
cellular protein metabolic process
|
0.686538 |
0.686538 |
1
|
1505
|
|
GO:0045449
|
regulation of transcription
|
0.687166 |
0.687166 |
1
|
771
|
|
GO:0031175
|
neuron projection development
|
0.696270 |
0.696270 |
1
|
392
|
|
GO:0003676
|
nucleic acid binding
|
0.697754 |
0.697754 |
1
|
1855
|
|
GO:0006909
|
phagocytosis
|
0.723404 |
0.723404 |
1
|
204
|
|
GO:0031224
|
intrinsic to membrane
|
0.725322 |
0.725322 |
1
|
1014
|
|
GO:0043234
|
protein complex
|
0.727104 |
0.727104 |
1
|
1564
|
|
GO:0004672
|
protein kinase activity
|
0.728643 |
0.728643 |
1
|
290
|
|
GO:0016773
|
phosphotransferase activity, alcohol group as acceptor
|
0.735354 |
0.735354 |
1
|
364
|
|
GO:0032990
|
cell part morphogenesis
|
0.745583 |
0.745583 |
1
|
422
|
|
GO:0032553
|
ribonucleotide binding
|
0.746180 |
0.746180 |
1
|
879
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.746994 |
0.746994 |
1
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.748407 |
0.748407 |
1
|
1959
|
|
GO:0003824
|
catalytic activity
|
0.751316 |
0.751316 |
1
|
4106
|
|
GO:0032559
|
adenyl ribonucleotide binding
|
0.757705 |
0.757705 |
1
|
713
|
|
GO:0016310
|
phosphorylation
|
0.758993 |
0.758993 |
1
|
422
|
|
GO:0043170
|
macromolecule metabolic process
|
0.772351 |
0.772351 |
2
|
3588
|
|
GO:0005575
|
cellular_component
|
0.773470 |
0.773470 |
2
|
8817
|
|
GO:0044249
|
cellular biosynthetic process
|
0.776153 |
0.776153 |
1
|
2034
|
|
GO:0010467
|
gene expression
|
0.780571 |
0.780571 |
1
|
1907
|
|
GO:0008270
|
zinc ion binding
|
0.781440 |
0.781440 |
1
|
901
|
|
GO:0009058
|
biosynthetic process
|
0.786651 |
0.786651 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.787269 |
0.787269 |
1
|
2093
|
|
GO:0048667
|
cell morphogenesis involved in neuron differentiation
|
0.795501 |
0.795501 |
1
|
389
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0016301
|
kinase activity
|
0.795960 |
0.795960 |
1
|
394
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.800673 |
0.800673 |
1
|
1789
|
|
GO:0017076
|
purine nucleotide binding
|
0.803056 |
0.803056 |
1
|
946
|
|
GO:0043229
|
intracellular organelle
|
0.804100 |
0.804100 |
1
|
3530
|
|
GO:0048858
|
cell projection morphogenesis
|
0.806883 |
0.806883 |
1
|
422
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.814170 |
0.814170 |
1
|
701
|
|
GO:0030554
|
adenyl nucleotide binding
|
0.819239 |
0.819239 |
1
|
775
|
|
GO:0010324
|
membrane invagination
|
0.819767 |
0.819767 |
1
|
282
|
|
GO:0004872
|
receptor activity
|
0.821109 |
0.821109 |
1
|
459
|
|
GO:0000902
|
cell morphogenesis
|
0.825088 |
0.825088 |
1
|
467
|
|
GO:0032934
|
sterol binding
|
0.833333 |
0.833333 |
1
|
10
|
|
GO:0019538
|
protein metabolic process
|
0.855761 |
0.855761 |
1
|
2053
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0004888
|
transmembrane receptor activity
|
0.860566 |
0.860566 |
1
|
395
|
|
GO:0048812
|
neuron projection morphogenesis
|
0.910798 |
0.910798 |
1
|
388
|
|
GO:0044424
|
intracellular part
|
0.914575 |
0.914575 |
1
|
4384
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.929175 |
0.929175 |
1
|
879
|
|
GO:0005622
|
intracellular
|
0.944411 |
0.944411 |
1
|
4734
|
|
GO:0016043
|
cellular component organization
|
0.947563 |
0.947563 |
2
|
2052
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0016021
|
integral to membrane
|
0.984221 |
0.984221 |
1
|
998
|
|
GO:0001883
|
purine nucleoside binding
|
0.992347 |
0.992347 |
1
|
778
|
|
GO:0005524
|
ATP binding
|
0.998597 |
0.998597 |
1
|
712
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0003700
|
sequence-specific DNA binding transcription factor activity
|
1.00000 |
1.00000 |
1
|
395
|
|
GO:0004871
|
signal transducer activity
|
1.00000 |
1.00000 |
1
|
559
|
|
GO:0006796
|
phosphate metabolic process
|
1.00000 |
1.00000 |
1
|
556
|