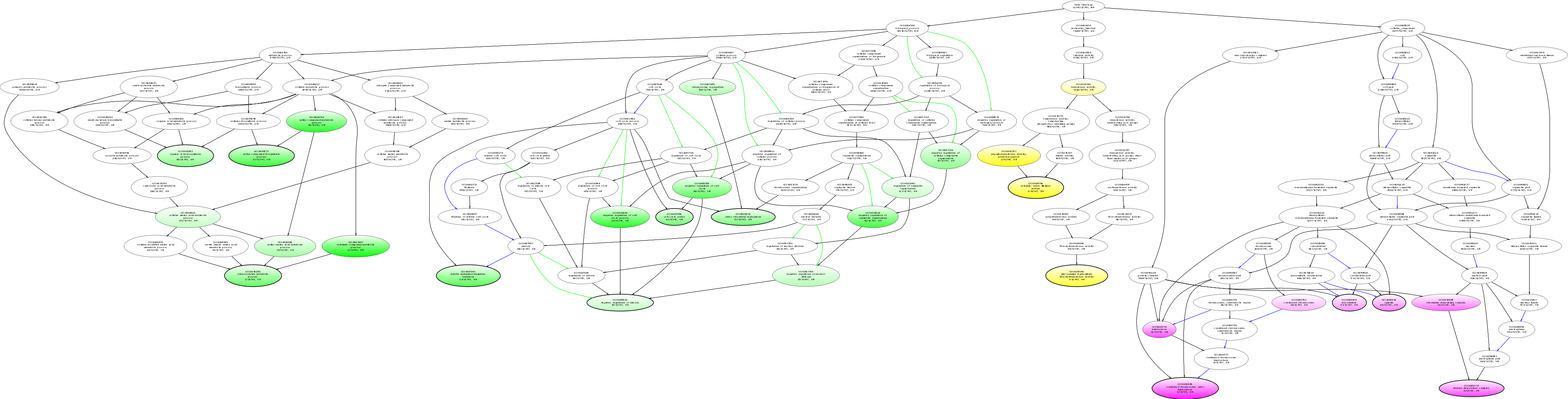
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0001887
|
selenium compound metabolic process
|
0.00104863 |
0.00104863 |
1
|
2
|
|
GO:0016781
|
phosphotransferase activity, paired acceptors
|
0.00404040 |
0.00404040 |
1
|
2
|
|
GO:0004756
|
selenide, water dikinase activity
|
0.00507614 |
0.00507614 |
1
|
2
|
|
GO:0000940
|
condensed chromosome outer kinetochore
|
0.00899630 |
0.00899630 |
1
|
8
|
|
GO:0010948
|
negative regulation of cell cycle process
|
0.0195246 |
0.0195246 |
1
|
23
|
|
GO:0006790
|
sulfur compound metabolic process
|
0.0198299 |
0.0198299 |
1
|
38
|
|
GO:0004343
|
glucosamine 6-phosphate N-acetyltransferase activity
|
0.0227273 |
0.0227273 |
1
|
1
|
|
GO:0044272
|
sulfur compound biosynthetic process
|
0.0262507 |
0.0262507 |
1
|
27
|
|
GO:0010639
|
negative regulation of organelle organization
|
0.0262515 |
0.0262515 |
1
|
43
|
|
GO:0000118
|
histone deacetylase complex
|
0.0277778 |
0.0277778 |
1
|
8
|
|
GO:0000776
|
kinetochore
|
0.0289643 |
0.0289643 |
1
|
32
|
|
GO:0007091
|
mitotic metaphase/anaphase transition
|
0.0351145 |
0.0351145 |
1
|
23
|
|
GO:0045786
|
negative regulation of cell cycle
|
0.0369181 |
0.0369181 |
1
|
46
|
|
GO:0016259
|
selenocysteine metabolic process
|
0.0377358 |
0.0377358 |
1
|
2
|
|
GO:0016585
|
chromatin remodeling complex
|
0.0380441 |
0.0380441 |
1
|
38
|
|
GO:0007050
|
cell cycle arrest
|
0.0502283 |
0.0502283 |
1
|
33
|
|
GO:0000819
|
sister chromatid segregation
|
0.0529595 |
0.0529595 |
1
|
51
|
|
GO:0005819
|
spindle
|
0.0554197 |
0.0554197 |
1
|
68
|
|
GO:0016053
|
organic acid biosynthetic process
|
0.0593473 |
0.0593473 |
1
|
65
|
|
GO:0051129
|
negative regulation of cellular component organization
|
0.0611847 |
0.0611847 |
1
|
75
|
|
GO:0005874
|
microtubule
|
0.0621796 |
0.0621796 |
1
|
51
|
|
GO:0016740
|
transferase activity
|
0.0628812 |
0.0628812 |
2
|
1030
|
|
GO:0000096
|
sulfur amino acid metabolic process
|
0.0736842 |
0.0736842 |
1
|
14
|
|
GO:0007059
|
chromosome segregation
|
0.0740208 |
0.0740208 |
1
|
166
|
|
GO:0000793
|
condensed chromosome
|
0.0761421 |
0.0761421 |
1
|
30
|
|
GO:0051784
|
negative regulation of nuclear division
|
0.0769231 |
0.0769231 |
1
|
15
|
|
GO:0045839
|
negative regulation of mitosis
|
0.0781250 |
0.0781250 |
1
|
15
|
|
GO:0006520
|
cellular amino acid metabolic process
|
0.0784268 |
0.0784268 |
1
|
172
|
|
GO:0033043
|
regulation of organelle organization
|
0.0890453 |
0.0890453 |
1
|
127
|
|
GO:0000097
|
sulfur amino acid biosynthetic process
|
0.100000 |
0.100000 |
1
|
6
|
|
GO:0009309
|
amine biosynthetic process
|
0.102119 |
0.102119 |
1
|
53
|
|
GO:0001071
|
nucleic acid binding transcription factor activity
|
0.103335 |
0.103335 |
1
|
395
|
|
GO:0009069
|
serine family amino acid metabolic process
|
0.104651 |
0.104651 |
1
|
18
|
|
GO:0016260
|
selenocysteine biosynthetic process
|
0.117647 |
0.117647 |
1
|
2
|
|
GO:0006082
|
organic acid metabolic process
|
0.130330 |
0.130330 |
1
|
259
|
|
GO:0042398
|
cellular modified amino acid biosynthetic process
|
0.140351 |
0.140351 |
1
|
8
|
|
GO:0042180
|
cellular ketone metabolic process
|
0.140499 |
0.140499 |
1
|
280
|
|
GO:0051726
|
regulation of cell cycle
|
0.144812 |
0.144812 |
1
|
187
|
|
GO:0010564
|
regulation of cell cycle process
|
0.148725 |
0.148725 |
1
|
105
|
|
GO:0048285
|
organelle fission
|
0.148900 |
0.148900 |
1
|
176
|
|
GO:0016044
|
cellular membrane organization
|
0.149233 |
0.149233 |
1
|
344
|
|
GO:0032991
|
macromolecular complex
|
0.149479 |
0.149479 |
2
|
2151
|
|
GO:0044422
|
organelle part
|
0.152629 |
0.152629 |
2
|
2176
|
|
GO:0007093
|
mitotic cell cycle checkpoint
|
0.156522 |
0.156522 |
1
|
18
|
|
GO:0051128
|
regulation of cellular component organization
|
0.156775 |
0.156775 |
1
|
282
|
|
GO:0006575
|
cellular modified amino acid metabolic process
|
0.162791 |
0.162791 |
1
|
28
|
|
GO:0000775
|
chromosome, centromeric region
|
0.163743 |
0.163743 |
1
|
56
|
|
GO:0000779
|
condensed chromosome, centromeric region
|
0.169014 |
0.169014 |
1
|
12
|
|
GO:0009070
|
serine family amino acid biosynthetic process
|
0.173913 |
0.173913 |
1
|
8
|
|
GO:0008652
|
cellular amino acid biosynthetic process
|
0.175355 |
0.175355 |
1
|
37
|
|
GO:0016192
|
vesicle-mediated transport
|
0.176723 |
0.176723 |
1
|
430
|
|
GO:0044271
|
cellular nitrogen compound biosynthetic process
|
0.177736 |
0.177736 |
1
|
252
|
|
GO:0044106
|
cellular amine metabolic process
|
0.178182 |
0.178182 |
1
|
195
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.178576 |
0.178576 |
2
|
2108
|
|
GO:0051783
|
regulation of nuclear division
|
0.185484 |
0.185484 |
1
|
46
|
|
GO:0007088
|
regulation of mitosis
|
0.187755 |
0.187755 |
1
|
46
|
|
GO:0050789
|
regulation of biological process
|
0.198789 |
0.198789 |
2
|
2246
|
|
GO:0044283
|
small molecule biosynthetic process
|
0.199369 |
0.199369 |
1
|
259
|
|
GO:0031974
|
membrane-enclosed lumen
|
0.204405 |
0.204405 |
1
|
647
|
|
GO:0007346
|
regulation of mitotic cell cycle
|
0.220165 |
0.220165 |
1
|
107
|
|
GO:0005876
|
spindle microtubule
|
0.221053 |
0.221053 |
1
|
21
|
|
GO:0050794
|
regulation of cellular process
|
0.224039 |
0.224039 |
2
|
2034
|
|
GO:0071156
|
regulation of cell cycle arrest
|
0.236364 |
0.236364 |
1
|
26
|
|
GO:0048519
|
negative regulation of biological process
|
0.240665 |
0.240665 |
1
|
706
|
|
GO:0044446
|
intracellular organelle part
|
0.243036 |
0.243036 |
2
|
2162
|
|
GO:0065007
|
biological regulation
|
0.244964 |
0.244964 |
2
|
2548
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.248286 |
0.248286 |
2
|
1959
|
|
GO:0046394
|
carboxylic acid biosynthetic process
|
0.250965 |
0.250965 |
1
|
65
|
|
GO:0016746
|
transferase activity, transferring acyl groups
|
0.253484 |
0.253484 |
1
|
140
|
|
GO:0001882
|
nucleoside binding
|
0.258670 |
0.258670 |
1
|
784
|
|
GO:0048523
|
negative regulation of cellular process
|
0.262869 |
0.262869 |
1
|
635
|
|
GO:0000777
|
condensed chromosome kinetochore
|
0.264706 |
0.264706 |
1
|
9
|
|
GO:0000087
|
M phase of mitotic cell cycle
|
0.269525 |
0.269525 |
1
|
176
|
|
GO:0022402
|
cell cycle process
|
0.270667 |
0.270667 |
1
|
655
|
|
GO:0044249
|
cellular biosynthetic process
|
0.277461 |
0.277461 |
2
|
2034
|
|
GO:0007067
|
mitosis
|
0.281356 |
0.281356 |
1
|
166
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.283034 |
0.283034 |
1
|
1056
|
|
GO:0009308
|
amine metabolic process
|
0.286510 |
0.286510 |
1
|
325
|
|
GO:0044427
|
chromosomal part
|
0.288122 |
0.288122 |
1
|
342
|
|
GO:0008152
|
metabolic process
|
0.296531 |
0.296531 |
3
|
5194
|
|
GO:0000070
|
mitotic sister chromatid segregation
|
0.299401 |
0.299401 |
1
|
50
|
|
GO:0043565
|
sequence-specific DNA binding
|
0.300971 |
0.300971 |
1
|
248
|
|
GO:0007049
|
cell cycle
|
0.307460 |
0.307460 |
1
|
756
|
|
GO:0061024
|
membrane organization
|
0.308058 |
0.308058 |
1
|
345
|
|
GO:0003824
|
catalytic activity
|
0.310936 |
0.310936 |
2
|
4106
|
|
GO:0044451
|
nucleoplasm part
|
0.313253 |
0.313253 |
1
|
260
|
|
GO:0005694
|
chromosome
|
0.321108 |
0.321108 |
1
|
394
|
|
GO:0005654
|
nucleoplasm
|
0.340964 |
0.340964 |
1
|
283
|
|
GO:0043226
|
organelle
|
0.353651 |
0.353651 |
2
|
3537
|
|
GO:0009058
|
biosynthetic process
|
0.355414 |
0.355414 |
2
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.356248 |
0.356248 |
2
|
2093
|
|
GO:0051276
|
chromosome organization
|
0.358714 |
0.358714 |
1
|
424
|
|
GO:0000166
|
nucleotide binding
|
0.374077 |
0.374077 |
1
|
1178
|
|
GO:0044281
|
small molecule metabolic process
|
0.378352 |
0.378352 |
1
|
761
|
|
GO:0006350
|
transcription
|
0.387810 |
0.387810 |
1
|
859
|
|
GO:0044430
|
cytoskeletal part
|
0.409756 |
0.409756 |
1
|
512
|
|
GO:0016070
|
RNA metabolic process
|
0.413829 |
0.413829 |
1
|
1191
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.418200 |
0.418200 |
1
|
1039
|
|
GO:0031577
|
spindle checkpoint
|
0.423077 |
0.423077 |
1
|
11
|
|
GO:0005828
|
kinetochore microtubule
|
0.428571 |
0.428571 |
1
|
9
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.440308 |
0.440308 |
1
|
686
|
|
GO:0003677
|
DNA binding
|
0.444205 |
0.444205 |
1
|
824
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.451726 |
0.451726 |
1
|
903
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.456989 |
0.456989 |
1
|
850
|
|
GO:0005856
|
cytoskeleton
|
0.457213 |
0.457213 |
1
|
561
|
|
GO:0016410
|
N-acyltransferase activity
|
0.460000 |
0.460000 |
1
|
46
|
|
GO:0043167
|
ion binding
|
0.460626 |
0.460626 |
1
|
1498
|
|
GO:0051234
|
establishment of localization
|
0.467930 |
0.467930 |
1
|
1549
|
|
GO:0044428
|
nuclear part
|
0.469235 |
0.469235 |
1
|
830
|
|
GO:0031981
|
nuclear lumen
|
0.475277 |
0.475277 |
1
|
471
|
|
GO:0008150
|
biological_process
|
0.481028 |
0.481028 |
4
|
10616
|
|
GO:0010468
|
regulation of gene expression
|
0.489930 |
0.489930 |
1
|
973
|
|
GO:0032774
|
RNA biosynthetic process
|
0.496008 |
0.496008 |
1
|
703
|
|
GO:0043233
|
organelle lumen
|
0.497925 |
0.497925 |
1
|
634
|
|
GO:0030071
|
regulation of mitotic metaphase/anaphase transition
|
0.500000 |
0.500000 |
1
|
23
|
|
GO:0070013
|
intracellular organelle lumen
|
0.500596 |
0.500596 |
1
|
634
|
|
GO:0044424
|
intracellular part
|
0.500760 |
0.500760 |
2
|
4384
|
|
GO:0009987
|
cellular process
|
0.506393 |
0.506393 |
3
|
6560
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.513595 |
0.513595 |
1
|
850
|
|
GO:0005488
|
binding
|
0.514777 |
0.514777 |
2
|
5641
|
|
GO:0045841
|
negative regulation of mitotic metaphase/anaphase transition
|
0.520000 |
0.520000 |
1
|
13
|
|
GO:0043234
|
protein complex
|
0.528587 |
0.528587 |
2
|
1564
|
|
GO:0010467
|
gene expression
|
0.531494 |
0.531494 |
1
|
1907
|
|
GO:0016407
|
acetyltransferase activity
|
0.540000 |
0.540000 |
1
|
54
|
|
GO:0051179
|
localization
|
0.544835 |
0.544835 |
1
|
1896
|
|
GO:0003676
|
nucleic acid binding
|
0.549587 |
0.549587 |
1
|
1855
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.556542 |
0.556542 |
1
|
1562
|
|
GO:0000278
|
mitotic cell cycle
|
0.567460 |
0.567460 |
1
|
429
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.569226 |
0.569226 |
1
|
1110
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.573530 |
0.573530 |
1
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.574428 |
0.574428 |
1
|
1227
|
|
GO:0005622
|
intracellular
|
0.583919 |
0.583919 |
2
|
4734
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.599142 |
0.599142 |
1
|
888
|
|
GO:0071174
|
mitotic cell cycle spindle checkpoint
|
0.611111 |
0.611111 |
1
|
11
|
|
GO:0019222
|
regulation of metabolic process
|
0.611396 |
0.611396 |
1
|
1242
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.617799 |
0.617799 |
1
|
905
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.635595 |
0.635595 |
1
|
888
|
|
GO:0005575
|
cellular_component
|
0.637024 |
0.637024 |
3
|
8817
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0043229
|
intracellular organelle
|
0.646541 |
0.646541 |
2
|
3530
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.646807 |
0.646807 |
1
|
1623
|
|
GO:0006897
|
endocytosis
|
0.655814 |
0.655814 |
1
|
282
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.658716 |
0.658716 |
1
|
1789
|
|
GO:0044237
|
cellular metabolic process
|
0.673909 |
0.673909 |
2
|
3814
|
|
GO:0006911
|
phagocytosis, engulfment
|
0.680851 |
0.680851 |
1
|
192
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.684327 |
0.684327 |
1
|
620
|
|
GO:0045449
|
regulation of transcription
|
0.687166 |
0.687166 |
1
|
771
|
|
GO:0006909
|
phagocytosis
|
0.723404 |
0.723404 |
1
|
204
|
|
GO:0016772
|
transferase activity, transferring phosphorus-containing groups
|
0.730448 |
0.730448 |
1
|
495
|
|
GO:0032553
|
ribonucleotide binding
|
0.746180 |
0.746180 |
1
|
879
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.746994 |
0.746994 |
1
|
1617
|
|
GO:0032559
|
adenyl ribonucleotide binding
|
0.757705 |
0.757705 |
1
|
713
|
|
GO:0006996
|
organelle organization
|
0.779169 |
0.779169 |
1
|
1182
|
|
GO:0008270
|
zinc ion binding
|
0.781440 |
0.781440 |
1
|
901
|
|
GO:0008080
|
N-acetyltransferase activity
|
0.785714 |
0.785714 |
1
|
44
|
|
GO:0005623
|
cell
|
0.787322 |
0.787322 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.787322 |
0.787322 |
2
|
6195
|
|
GO:0000075
|
cell cycle checkpoint
|
0.787879 |
0.787879 |
1
|
26
|
|
GO:0005737
|
cytoplasm
|
0.790684 |
0.790684 |
1
|
2378
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0016301
|
kinase activity
|
0.795960 |
0.795960 |
1
|
394
|
|
GO:0015630
|
microtubule cytoskeleton
|
0.798574 |
0.798574 |
1
|
448
|
|
GO:0017076
|
purine nucleotide binding
|
0.803056 |
0.803056 |
1
|
946
|
|
GO:0008415
|
acyltransferase activity
|
0.813008 |
0.813008 |
1
|
100
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.814170 |
0.814170 |
1
|
701
|
|
GO:0030554
|
adenyl nucleotide binding
|
0.819239 |
0.819239 |
1
|
775
|
|
GO:0010324
|
membrane invagination
|
0.819767 |
0.819767 |
1
|
282
|
|
GO:0007094
|
mitotic cell cycle spindle assembly checkpoint
|
0.846154 |
0.846154 |
1
|
11
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.869824 |
0.869824 |
1
|
2878
|
|
GO:0016747
|
transferase activity, transferring acyl groups other than amino-acyl groups
|
0.878571 |
0.878571 |
1
|
123
|
|
GO:0022403
|
cell cycle phase
|
0.896183 |
0.896183 |
1
|
587
|
|
GO:0003674
|
molecular_function
|
0.912936 |
0.912936 |
3
|
11064
|
|
GO:0044238
|
primary metabolic process
|
0.914505 |
0.914505 |
2
|
4259
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.923977 |
0.923977 |
1
|
1517
|
|
GO:0043436
|
oxoacid metabolic process
|
0.925000 |
0.925000 |
1
|
259
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.929175 |
0.929175 |
1
|
879
|
|
GO:0000279
|
M phase
|
0.945486 |
0.945486 |
1
|
555
|
|
GO:0016043
|
cellular component organization
|
0.947563 |
0.947563 |
2
|
2052
|
|
GO:0043227
|
membrane-bounded organelle
|
0.963300 |
0.963300 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.963413 |
0.963413 |
1
|
2856
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0043170
|
macromolecule metabolic process
|
0.970476 |
0.970476 |
1
|
3588
|
|
GO:0000280
|
nuclear division
|
0.971591 |
0.971591 |
1
|
171
|
|
GO:0001883
|
purine nucleoside binding
|
0.992347 |
0.992347 |
1
|
778
|
|
GO:0005524
|
ATP binding
|
0.998597 |
0.998597 |
1
|
712
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
4
|
12747
|
|
GO:0071173
|
spindle assembly checkpoint
|
1.00000 |
1.00000 |
1
|
11
|
|
GO:0019752
|
carboxylic acid metabolic process
|
1.00000 |
1.00000 |
1
|
259
|
|
GO:0003700
|
sequence-specific DNA binding transcription factor activity
|
1.00000 |
1.00000 |
1
|
395
|