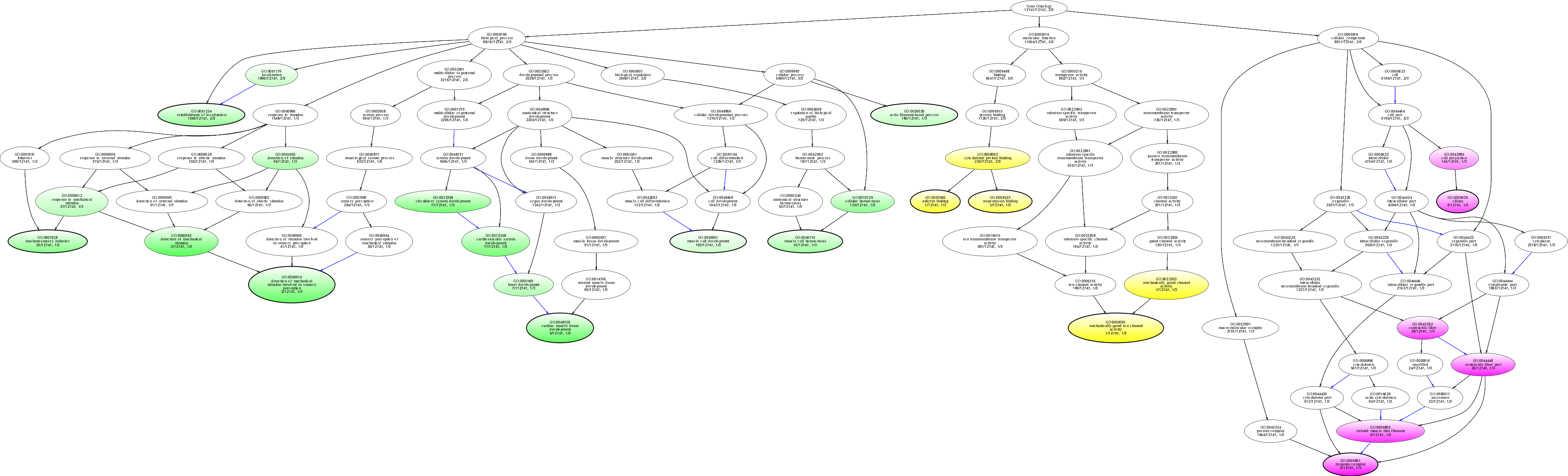
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0005861
|
troponin complex
|
0.00123305 |
0.00123305 |
1
|
2
|
|
GO:0008381
|
mechanically-gated ion channel activity
|
0.00529101 |
0.00529101 |
1
|
1
|
|
GO:0022833
|
mechanically gated channel activity
|
0.00769231 |
0.00769231 |
1
|
1
|
|
GO:0044449
|
contractile fiber part
|
0.00848010 |
0.00848010 |
1
|
26
|
|
GO:0030506
|
ankyrin binding
|
0.00909091 |
0.00909091 |
1
|
1
|
|
GO:0005865
|
striated muscle thin filament
|
0.00934579 |
0.00934579 |
1
|
5
|
|
GO:0043292
|
contractile fiber
|
0.0107157 |
0.0107157 |
1
|
28
|
|
GO:0008092
|
cytoskeletal protein binding
|
0.0161822 |
0.0161822 |
2
|
220
|
|
GO:0005929
|
cilium
|
0.0209790 |
0.0209790 |
1
|
3
|
|
GO:0005523
|
tropomyosin binding
|
0.0271482 |
0.0271482 |
1
|
3
|
|
GO:0050982
|
detection of mechanical stimulus
|
0.0315789 |
0.0315789 |
1
|
3
|
|
GO:0048738
|
cardiac muscle tissue development
|
0.0331126 |
0.0331126 |
1
|
5
|
|
GO:0050974
|
detection of mechanical stimulus involved in sensory perception
|
0.0405405 |
0.0405405 |
1
|
3
|
|
GO:0072359
|
circulatory system development
|
0.0454009 |
0.0454009 |
1
|
77
|
|
GO:0072358
|
cardiovascular system development
|
0.0454009 |
0.0454009 |
1
|
77
|
|
GO:0042995
|
cell projection
|
0.0456371 |
0.0456371 |
1
|
143
|
|
GO:0007638
|
mechanosensory behavior
|
0.0462633 |
0.0462633 |
1
|
26
|
|
GO:0019725
|
cellular homeostasis
|
0.0560655 |
0.0560655 |
1
|
125
|
|
GO:0051234
|
establishment of localization
|
0.0576328 |
0.0576328 |
2
|
1549
|
|
GO:0046716
|
muscle cell homeostasis
|
0.0602410 |
0.0602410 |
1
|
10
|
|
GO:0051606
|
detection of stimulus
|
0.0613695 |
0.0613695 |
1
|
95
|
|
GO:0007507
|
heart development
|
0.0619968 |
0.0619968 |
1
|
77
|
|
GO:0009612
|
response to mechanical stimulus
|
0.0756646 |
0.0756646 |
1
|
37
|
|
GO:0030029
|
actin filament-based process
|
0.0826842 |
0.0826842 |
1
|
186
|
|
GO:0051179
|
localization
|
0.0842718 |
0.0842718 |
2
|
1896
|
|
GO:0055001
|
muscle cell development
|
0.0938967 |
0.0938967 |
1
|
100
|
|
GO:0042692
|
muscle cell differentiation
|
0.102644 |
0.102644 |
1
|
132
|
|
GO:0061061
|
muscle structure development
|
0.111258 |
0.111258 |
1
|
252
|
|
GO:0030239
|
myofibril assembly
|
0.117886 |
0.117886 |
1
|
29
|
|
GO:0050906
|
detection of stimulus involved in sensory perception
|
0.125767 |
0.125767 |
1
|
41
|
|
GO:0050954
|
sensory perception of mechanical stimulus
|
0.126761 |
0.126761 |
1
|
36
|
|
GO:0005488
|
binding
|
0.132501 |
0.132501 |
3
|
5641
|
|
GO:0010927
|
cellular component assembly involved in morphogenesis
|
0.134157 |
0.134157 |
1
|
152
|
|
GO:0065007
|
biological regulation
|
0.145142 |
0.145142 |
2
|
2548
|
|
GO:0048285
|
organelle fission
|
0.148900 |
0.148900 |
1
|
176
|
|
GO:0007517
|
muscle organ development
|
0.149102 |
0.149102 |
1
|
191
|
|
GO:0009581
|
detection of external stimulus
|
0.149510 |
0.149510 |
1
|
61
|
|
GO:0072511
|
divalent inorganic cation transport
|
0.149798 |
0.149798 |
1
|
37
|
|
GO:0060537
|
muscle tissue development
|
0.163375 |
0.163375 |
1
|
91
|
|
GO:0009628
|
response to abiotic stimulus
|
0.164729 |
0.164729 |
1
|
255
|
|
GO:0015629
|
actin cytoskeleton
|
0.167558 |
0.167558 |
1
|
94
|
|
GO:0048646
|
anatomical structure formation involved in morphogenesis
|
0.176649 |
0.176649 |
1
|
466
|
|
GO:0019001
|
guanyl nucleotide binding
|
0.182875 |
0.182875 |
1
|
173
|
|
GO:0009582
|
detection of abiotic stimulus
|
0.190476 |
0.190476 |
1
|
56
|
|
GO:0055085
|
transmembrane transport
|
0.191633 |
0.191633 |
1
|
469
|
|
GO:0032989
|
cellular component morphogenesis
|
0.193108 |
0.193108 |
1
|
566
|
|
GO:0032561
|
guanyl ribonucleotide binding
|
0.194098 |
0.194098 |
1
|
171
|
|
GO:0023046
|
signaling process
|
0.199779 |
0.199779 |
1
|
760
|
|
GO:0005215
|
transporter activity
|
0.213704 |
0.213704 |
1
|
852
|
|
GO:0005262
|
calcium channel activity
|
0.218750 |
0.218750 |
1
|
28
|
|
GO:0005515
|
protein binding
|
0.223526 |
0.223526 |
2
|
1726
|
|
GO:0070838
|
divalent metal ion transport
|
0.227848 |
0.227848 |
1
|
36
|
|
GO:0032501
|
multicellular organismal process
|
0.231607 |
0.231607 |
2
|
3315
|
|
GO:0044430
|
cytoskeletal part
|
0.231674 |
0.231674 |
1
|
512
|
|
GO:0009987
|
cellular process
|
0.235914 |
0.235914 |
3
|
6560
|
|
GO:0003924
|
GTPase activity
|
0.241796 |
0.241796 |
1
|
140
|
|
GO:0009605
|
response to external stimulus
|
0.242248 |
0.242248 |
1
|
375
|
|
GO:0009888
|
tissue development
|
0.245916 |
0.245916 |
1
|
557
|
|
GO:0007165
|
signal transduction
|
0.254057 |
0.254057 |
1
|
548
|
|
GO:0042592
|
homeostatic process
|
0.256516 |
0.256516 |
1
|
187
|
|
GO:0005261
|
cation channel activity
|
0.272340 |
0.272340 |
1
|
128
|
|
GO:0022607
|
cellular component assembly
|
0.275418 |
0.275418 |
1
|
577
|
|
GO:0022803
|
passive transmembrane transporter activity
|
0.276860 |
0.276860 |
1
|
201
|
|
GO:0060249
|
anatomical structure homeostasis
|
0.283422 |
0.283422 |
1
|
53
|
|
GO:0044085
|
cellular component biogenesis
|
0.299810 |
0.299810 |
1
|
632
|
|
GO:0016817
|
hydrolase activity, acting on acid anhydrides
|
0.301217 |
0.301217 |
1
|
594
|
|
GO:0031032
|
actomyosin structure organization
|
0.306011 |
0.306011 |
1
|
56
|
|
GO:0022838
|
substrate-specific channel activity
|
0.307448 |
0.307448 |
1
|
194
|
|
GO:0045184
|
establishment of protein localization
|
0.308898 |
0.308898 |
1
|
273
|
|
GO:0015031
|
protein transport
|
0.323317 |
0.323317 |
1
|
266
|
|
GO:0023052
|
signaling
|
0.338079 |
0.338079 |
1
|
1364
|
|
GO:0030036
|
actin cytoskeleton organization
|
0.339518 |
0.339518 |
1
|
183
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.346904 |
0.346904 |
1
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.347592 |
0.347592 |
1
|
1227
|
|
GO:0045214
|
sarcomere organization
|
0.357143 |
0.357143 |
1
|
20
|
|
GO:0003008
|
system process
|
0.360531 |
0.360531 |
1
|
664
|
|
GO:0007610
|
behavior
|
0.361111 |
0.361111 |
1
|
559
|
|
GO:0005216
|
ion channel activity
|
0.369141 |
0.369141 |
1
|
189
|
|
GO:0071844
|
cellular component assembly at cellular level
|
0.373193 |
0.373193 |
1
|
568
|
|
GO:0050896
|
response to stimulus
|
0.376795 |
0.376795 |
1
|
1548
|
|
GO:0006811
|
ion transport
|
0.393444 |
0.393444 |
1
|
331
|
|
GO:0044444
|
cytoplasmic part
|
0.424954 |
0.424954 |
1
|
1863
|
|
GO:0032991
|
macromolecular complex
|
0.428425 |
0.428425 |
1
|
2151
|
|
GO:0023034
|
intracellular signaling pathway
|
0.430962 |
0.430962 |
1
|
412
|
|
GO:0044422
|
organelle part
|
0.432705 |
0.432705 |
1
|
2176
|
|
GO:0048869
|
cellular developmental process
|
0.442161 |
0.442161 |
1
|
1276
|
|
GO:0007600
|
sensory perception
|
0.448657 |
0.448657 |
1
|
284
|
|
GO:0007010
|
cytoskeleton organization
|
0.453469 |
0.453469 |
1
|
536
|
|
GO:0005856
|
cytoskeleton
|
0.457213 |
0.457213 |
1
|
561
|
|
GO:0048468
|
cell development
|
0.457419 |
0.457419 |
1
|
1042
|
|
GO:0033036
|
macromolecule localization
|
0.476467 |
0.476467 |
1
|
524
|
|
GO:0016787
|
hydrolase activity
|
0.480273 |
0.480273 |
1
|
1972
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.485282 |
0.485282 |
1
|
2108
|
|
GO:0065008
|
regulation of biological quality
|
0.490437 |
0.490437 |
1
|
729
|
|
GO:0044446
|
intracellular organelle part
|
0.493044 |
0.493044 |
1
|
2162
|
|
GO:0005623
|
cell
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0035556
|
intracellular signal transduction
|
0.499208 |
0.499208 |
1
|
315
|
|
GO:0007399
|
nervous system development
|
0.500000 |
0.500000 |
1
|
848
|
|
GO:0000166
|
nucleotide binding
|
0.504833 |
0.504833 |
1
|
1178
|
|
GO:0050789
|
regulation of biological process
|
0.509927 |
0.509927 |
1
|
2246
|
|
GO:0060538
|
skeletal muscle organ development
|
0.539267 |
0.539267 |
1
|
103
|
|
GO:0005737
|
cytoplasm
|
0.542427 |
0.542427 |
1
|
2378
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.556542 |
0.556542 |
1
|
1562
|
|
GO:0016020
|
membrane
|
0.567775 |
0.567775 |
1
|
2122
|
|
GO:0032502
|
developmental process
|
0.575616 |
0.575616 |
1
|
2638
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0007264
|
small GTPase mediated signal transduction
|
0.587302 |
0.587302 |
1
|
185
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0030001
|
metal ion transport
|
0.639676 |
0.639676 |
1
|
158
|
|
GO:0043226
|
organelle
|
0.641414 |
0.641414 |
1
|
3537
|
|
GO:0022836
|
gated channel activity
|
0.646766 |
0.646766 |
1
|
130
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0050794
|
regulation of cellular process
|
0.666348 |
0.666348 |
1
|
2034
|
|
GO:0007519
|
skeletal muscle tissue development
|
0.669565 |
0.669565 |
1
|
77
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.724105 |
0.724105 |
1
|
1517
|
|
GO:0008104
|
protein localization
|
0.727099 |
0.727099 |
1
|
381
|
|
GO:0043234
|
protein complex
|
0.727104 |
0.727104 |
1
|
1564
|
|
GO:0032553
|
ribonucleotide binding
|
0.746180 |
0.746180 |
1
|
879
|
|
GO:0006812
|
cation transport
|
0.746224 |
0.746224 |
1
|
247
|
|
GO:0003824
|
catalytic activity
|
0.751316 |
0.751316 |
1
|
4106
|
|
GO:0005575
|
cellular_component
|
0.773470 |
0.773470 |
2
|
8817
|
|
GO:0006996
|
organelle organization
|
0.779169 |
0.779169 |
1
|
1182
|
|
GO:0022891
|
substrate-specific transmembrane transporter activity
|
0.788146 |
0.788146 |
1
|
625
|
|
GO:0003779
|
actin binding
|
0.794521 |
0.794521 |
1
|
120
|
|
GO:0055002
|
striated muscle cell development
|
0.798387 |
0.798387 |
1
|
99
|
|
GO:0017076
|
purine nucleotide binding
|
0.803056 |
0.803056 |
1
|
946
|
|
GO:0043229
|
intracellular organelle
|
0.804100 |
0.804100 |
1
|
3530
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0008324
|
cation transmembrane transporter activity
|
0.808679 |
0.808679 |
1
|
410
|
|
GO:0015075
|
ion transmembrane transporter activity
|
0.811200 |
0.811200 |
1
|
507
|
|
GO:0022892
|
substrate-specific transporter activity
|
0.815728 |
0.815728 |
1
|
695
|
|
GO:0007605
|
sensory perception of sound
|
0.833333 |
0.833333 |
1
|
30
|
|
GO:0022857
|
transmembrane transporter activity
|
0.852113 |
0.852113 |
1
|
726
|
|
GO:0030016
|
myofibril
|
0.857143 |
0.857143 |
1
|
24
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0030017
|
sarcomere
|
0.884615 |
0.884615 |
1
|
23
|
|
GO:0007275
|
multicellular organismal development
|
0.887792 |
0.887792 |
1
|
2296
|
|
GO:0006816
|
calcium ion transport
|
0.888889 |
0.888889 |
1
|
32
|
|
GO:0044424
|
intracellular part
|
0.914575 |
0.914575 |
1
|
4384
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.929175 |
0.929175 |
1
|
879
|
|
GO:0006810
|
transport
|
0.933966 |
0.933966 |
2
|
1497
|
|
GO:0051146
|
striated muscle cell differentiation
|
0.939394 |
0.939394 |
1
|
124
|
|
GO:0005622
|
intracellular
|
0.944411 |
0.944411 |
1
|
4734
|
|
GO:0050877
|
neurological system process
|
0.953313 |
0.953313 |
1
|
633
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0000280
|
nuclear division
|
0.971591 |
0.971591 |
1
|
171
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0017111
|
nucleoside-triphosphatase activity
|
0.983022 |
0.983022 |
1
|
579
|
|
GO:0014706
|
striated muscle tissue development
|
0.989011 |
0.989011 |
1
|
90
|
|
GO:0016462
|
pyrophosphatase activity
|
0.991582 |
0.991582 |
1
|
589
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0016818
|
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
|
1.00000 |
1.00000 |
1
|
594
|
|
GO:0005525
|
GTP binding
|
1.00000 |
1.00000 |
1
|
171
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|
|
GO:0015267
|
channel activity
|
1.00000 |
1.00000 |
1
|
201
|