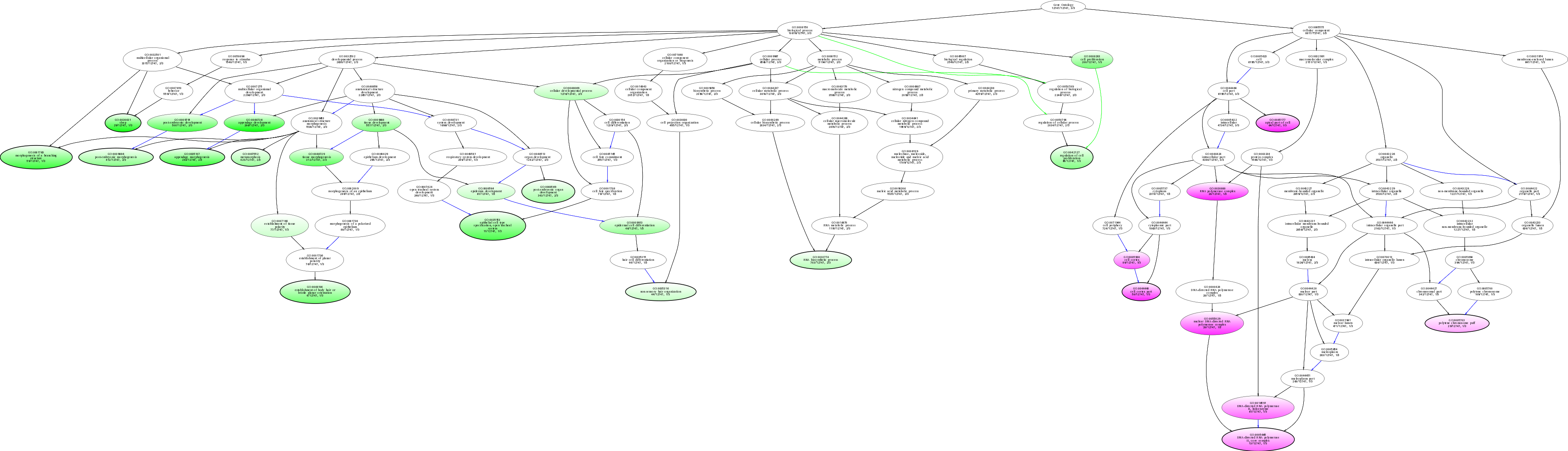
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0048736
|
appendage development
|
0.0122596 |
0.0122596 |
2
|
286
|
|
GO:0030431
|
sleep
|
0.0168749 |
0.0168749 |
1
|
29
|
|
GO:0030880
|
RNA polymerase complex
|
0.0182684 |
0.0182684 |
1
|
28
|
|
GO:0045177
|
apical part of cell
|
0.0306786 |
0.0306786 |
1
|
64
|
|
GO:0044448
|
cell cortex part
|
0.0311326 |
0.0311326 |
1
|
58
|
|
GO:0055029
|
nuclear DNA-directed RNA polymerase complex
|
0.0337349 |
0.0337349 |
1
|
28
|
|
GO:0035107
|
appendage morphogenesis
|
0.0338924 |
0.0338924 |
2
|
283
|
|
GO:0001763
|
morphogenesis of a branching structure
|
0.0347851 |
0.0347851 |
1
|
59
|
|
GO:0009791
|
post-embryonic development
|
0.0358662 |
0.0358662 |
2
|
500
|
|
GO:0035153
|
epithelial cell type specification, open tracheal system
|
0.0361842 |
0.0361842 |
1
|
11
|
|
GO:0005938
|
cell cortex
|
0.0369141 |
0.0369141 |
1
|
89
|
|
GO:0048729
|
tissue morphogenesis
|
0.0375970 |
0.0375970 |
2
|
312
|
|
GO:0016591
|
DNA-directed RNA polymerase II, holoenzyme
|
0.0378177 |
0.0378177 |
1
|
61
|
|
GO:0005665
|
DNA-directed RNA polymerase II, core complex
|
0.0490566 |
0.0490566 |
1
|
13
|
|
GO:0048104
|
establishment of body hair or bristle planar orientation
|
0.0526316 |
0.0526316 |
1
|
4
|
|
GO:0008283
|
cell proliferation
|
0.0554655 |
0.0554655 |
1
|
200
|
|
GO:0009888
|
tissue development
|
0.0603928 |
0.0603928 |
2
|
557
|
|
GO:0042127
|
regulation of cell proliferation
|
0.0612481 |
0.0612481 |
1
|
65
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.0671911 |
0.0671911 |
2
|
412
|
|
GO:0009913
|
epidermal cell differentiation
|
0.0726163 |
0.0726163 |
1
|
46
|
|
GO:0007552
|
metamorphosis
|
0.0748335 |
0.0748335 |
2
|
420
|
|
GO:0048569
|
post-embryonic organ development
|
0.0761074 |
0.0761074 |
2
|
343
|
|
GO:0005703
|
polytene chromosome puff
|
0.0773333 |
0.0773333 |
1
|
29
|
|
GO:0048869
|
cellular developmental process
|
0.0826655 |
0.0826655 |
2
|
1276
|
|
GO:0032774
|
RNA biosynthetic process
|
0.0840246 |
0.0840246 |
2
|
703
|
|
GO:0008544
|
epidermis development
|
0.0858137 |
0.0858137 |
1
|
61
|
|
GO:0035316
|
non-sensory hair organization
|
0.0907216 |
0.0907216 |
1
|
44
|
|
GO:0007164
|
establishment of tissue polarity
|
0.0979026 |
0.0979026 |
1
|
77
|
|
GO:0035295
|
tube development
|
0.100500 |
0.100500 |
1
|
133
|
|
GO:0001737
|
establishment of imaginal disc-derived wing hair orientation
|
0.102607 |
0.102607 |
1
|
23
|
|
GO:0001071
|
nucleic acid binding transcription factor activity
|
0.103335 |
0.103335 |
1
|
395
|
|
GO:0030528
|
transcription regulator activity
|
0.112632 |
0.112632 |
1
|
432
|
|
GO:0050789
|
regulation of biological process
|
0.115315 |
0.115315 |
2
|
2246
|
|
GO:0001708
|
cell fate specification
|
0.120037 |
0.120037 |
1
|
79
|
|
GO:0005488
|
binding
|
0.132501 |
0.132501 |
3
|
5641
|
|
GO:0009887
|
organ morphogenesis
|
0.140035 |
0.140035 |
2
|
684
|
|
GO:0007444
|
imaginal disc development
|
0.141192 |
0.141192 |
2
|
467
|
|
GO:0065007
|
biological regulation
|
0.145142 |
0.145142 |
2
|
2548
|
|
GO:0035239
|
tube morphogenesis
|
0.146222 |
0.146222 |
1
|
117
|
|
GO:0006350
|
transcription
|
0.150290 |
0.150290 |
2
|
859
|
|
GO:0007475
|
apposition of dorsal and ventral imaginal disc-derived wing surfaces
|
0.153958 |
0.153958 |
1
|
33
|
|
GO:0032502
|
developmental process
|
0.154531 |
0.154531 |
2
|
2638
|
|
GO:0044427
|
chromosomal part
|
0.156236 |
0.156236 |
1
|
342
|
|
GO:0009058
|
biosynthetic process
|
0.161869 |
0.161869 |
2
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.162334 |
0.162334 |
2
|
2093
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.164491 |
0.164491 |
2
|
1623
|
|
GO:0016779
|
nucleotidyltransferase activity
|
0.169697 |
0.169697 |
1
|
84
|
|
GO:0016070
|
RNA metabolic process
|
0.171170 |
0.171170 |
2
|
1191
|
|
GO:0048563
|
post-embryonic organ morphogenesis
|
0.172051 |
0.172051 |
2
|
323
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.172797 |
0.172797 |
2
|
1789
|
|
GO:0003677
|
DNA binding
|
0.197185 |
0.197185 |
2
|
824
|
|
GO:0031974
|
membrane-enclosed lumen
|
0.204405 |
0.204405 |
1
|
647
|
|
GO:0030030
|
cell projection organization
|
0.205631 |
0.205631 |
1
|
485
|
|
GO:0060446
|
branching involved in open tracheal system development
|
0.208178 |
0.208178 |
1
|
56
|
|
GO:0007478
|
leg disc morphogenesis
|
0.211371 |
0.211371 |
1
|
37
|
|
GO:0050794
|
regulation of cellular process
|
0.224039 |
0.224039 |
2
|
2034
|
|
GO:0035218
|
leg disc development
|
0.229444 |
0.229444 |
1
|
57
|
|
GO:0032501
|
multicellular organismal process
|
0.231607 |
0.231607 |
2
|
3315
|
|
GO:0009987
|
cellular process
|
0.235914 |
0.235914 |
3
|
6560
|
|
GO:0035127
|
post-embryonic limb morphogenesis
|
0.240385 |
0.240385 |
1
|
35
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.246859 |
0.246859 |
2
|
1617
|
|
GO:0035109
|
imaginal disc-derived limb morphogenesis
|
0.247235 |
0.247235 |
1
|
37
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.248286 |
0.248286 |
2
|
1959
|
|
GO:0016740
|
transferase activity
|
0.250852 |
0.250852 |
1
|
1030
|
|
GO:0003676
|
nucleic acid binding
|
0.253252 |
0.253252 |
2
|
1855
|
|
GO:0060541
|
respiratory system development
|
0.272151 |
0.272151 |
1
|
249
|
|
GO:0005700
|
polytene chromosome
|
0.276650 |
0.276650 |
1
|
109
|
|
GO:0044249
|
cellular biosynthetic process
|
0.277461 |
0.277461 |
2
|
2034
|
|
GO:0060429
|
epithelium development
|
0.280052 |
0.280052 |
2
|
295
|
|
GO:0010467
|
gene expression
|
0.282416 |
0.282416 |
2
|
1907
|
|
GO:0008587
|
imaginal disc-derived wing margin morphogenesis
|
0.286868 |
0.286868 |
1
|
64
|
|
GO:0043233
|
organelle lumen
|
0.291360 |
0.291360 |
1
|
634
|
|
GO:0070013
|
intracellular organelle lumen
|
0.293247 |
0.293247 |
1
|
634
|
|
GO:0048513
|
organ development
|
0.300572 |
0.300572 |
2
|
1242
|
|
GO:0060173
|
limb development
|
0.302147 |
0.302147 |
1
|
47
|
|
GO:0035108
|
limb morphogenesis
|
0.305065 |
0.305065 |
1
|
47
|
|
GO:0035317
|
imaginal disc-derived wing hair organization
|
0.309372 |
0.309372 |
1
|
43
|
|
GO:0071944
|
cell periphery
|
0.311271 |
0.311271 |
1
|
724
|
|
GO:0044451
|
nucleoplasm part
|
0.313253 |
0.313253 |
1
|
260
|
|
GO:0005694
|
chromosome
|
0.321108 |
0.321108 |
1
|
394
|
|
GO:0045179
|
apical cortex
|
0.322222 |
0.322222 |
1
|
29
|
|
GO:0005575
|
cellular_component
|
0.330897 |
0.330897 |
3
|
8817
|
|
GO:0007560
|
imaginal disc morphogenesis
|
0.331061 |
0.331061 |
2
|
323
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.338051 |
0.338051 |
2
|
1534
|
|
GO:0005654
|
nucleoplasm
|
0.340964 |
0.340964 |
1
|
283
|
|
GO:0005623
|
cell
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0044464
|
cell part
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0061138
|
morphogenesis of a branching epithelium
|
0.348383 |
0.348383 |
1
|
56
|
|
GO:0043226
|
organelle
|
0.353651 |
0.353651 |
2
|
3537
|
|
GO:0044424
|
intracellular part
|
0.354324 |
0.354324 |
3
|
4384
|
|
GO:0045165
|
cell fate commitment
|
0.359875 |
0.359875 |
1
|
255
|
|
GO:0007610
|
behavior
|
0.361111 |
0.361111 |
1
|
559
|
|
GO:0007389
|
pattern specification process
|
0.365750 |
0.365750 |
1
|
537
|
|
GO:0007430
|
terminal branching, open tracheal system
|
0.375000 |
0.375000 |
1
|
21
|
|
GO:0050896
|
response to stimulus
|
0.376795 |
0.376795 |
1
|
1548
|
|
GO:0035120
|
post-embryonic appendage morphogenesis
|
0.406614 |
0.406614 |
2
|
268
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.408435 |
0.408435 |
2
|
2878
|
|
GO:0005634
|
nucleus
|
0.408695 |
0.408695 |
2
|
1826
|
|
GO:0034062
|
RNA polymerase activity
|
0.416667 |
0.416667 |
1
|
35
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.418200 |
0.418200 |
1
|
1039
|
|
GO:0048731
|
system development
|
0.432376 |
0.432376 |
2
|
1696
|
|
GO:0007275
|
multicellular organismal development
|
0.442066 |
0.442066 |
2
|
2296
|
|
GO:0007472
|
wing disc morphogenesis
|
0.442702 |
0.442702 |
2
|
255
|
|
GO:0005622
|
intracellular
|
0.446165 |
0.446165 |
3
|
4734
|
|
GO:0035154
|
terminal cell fate specification, open tracheal system
|
0.454545 |
0.454545 |
1
|
5
|
|
GO:0035220
|
wing disc development
|
0.463216 |
0.463216 |
2
|
318
|
|
GO:0048754
|
branching morphogenesis of a tube
|
0.466667 |
0.466667 |
1
|
56
|
|
GO:0044428
|
nuclear part
|
0.469235 |
0.469235 |
1
|
830
|
|
GO:0031981
|
nuclear lumen
|
0.475277 |
0.475277 |
1
|
471
|
|
GO:0043170
|
macromolecule metabolic process
|
0.477159 |
0.477159 |
2
|
3588
|
|
GO:0016772
|
transferase activity, transferring phosphorus-containing groups
|
0.480583 |
0.480583 |
1
|
495
|
|
GO:0008152
|
metabolic process
|
0.483893 |
0.483893 |
2
|
5194
|
|
GO:0060562
|
epithelial tube morphogenesis
|
0.483903 |
0.483903 |
1
|
88
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.485282 |
0.485282 |
1
|
2108
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.486014 |
0.486014 |
1
|
1056
|
|
GO:0044237
|
cellular metabolic process
|
0.486483 |
0.486483 |
2
|
3814
|
|
GO:0019222
|
regulation of metabolic process
|
0.507779 |
0.507779 |
1
|
1242
|
|
GO:0043565
|
sequence-specific DNA binding
|
0.511614 |
0.511614 |
1
|
248
|
|
GO:0001738
|
morphogenesis of a polarized epithelium
|
0.517013 |
0.517013 |
1
|
88
|
|
GO:0032991
|
macromolecular complex
|
0.567899 |
0.567899 |
1
|
2151
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.569226 |
0.569226 |
1
|
1110
|
|
GO:0044422
|
organelle part
|
0.572743 |
0.572743 |
1
|
2176
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.573530 |
0.573530 |
1
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.574428 |
0.574428 |
1
|
1227
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0006366
|
transcription from RNA polymerase II promoter
|
0.591897 |
0.591897 |
1
|
253
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.599142 |
0.599142 |
1
|
888
|
|
GO:0043167
|
ion binding
|
0.603911 |
0.603911 |
1
|
1498
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.617799 |
0.617799 |
1
|
905
|
|
GO:0003702
|
RNA polymerase II transcription factor activity
|
0.618056 |
0.618056 |
1
|
267
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.635595 |
0.635595 |
1
|
888
|
|
GO:0043227
|
membrane-bounded organelle
|
0.653325 |
0.653325 |
2
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.653801 |
0.653801 |
2
|
2856
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.662696 |
0.662696 |
2
|
701
|
|
GO:0044238
|
primary metabolic process
|
0.672346 |
0.672346 |
2
|
4259
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.686903 |
0.686903 |
1
|
686
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.699519 |
0.699519 |
1
|
903
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.705273 |
0.705273 |
1
|
850
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.709978 |
0.709978 |
2
|
402
|
|
GO:0043234
|
protein complex
|
0.727104 |
0.727104 |
1
|
1564
|
|
GO:0007480
|
imaginal disc-derived leg morphogenesis
|
0.729167 |
0.729167 |
1
|
35
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.736132 |
0.736132 |
2
|
1535
|
|
GO:0048856
|
anatomical structure development
|
0.737157 |
0.737157 |
2
|
2265
|
|
GO:0010468
|
regulation of gene expression
|
0.739954 |
0.739954 |
1
|
973
|
|
GO:0003824
|
catalytic activity
|
0.751316 |
0.751316 |
1
|
4106
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.763561 |
0.763561 |
1
|
850
|
|
GO:0008270
|
zinc ion binding
|
0.781440 |
0.781440 |
1
|
901
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0044444
|
cytoplasmic part
|
0.809942 |
0.809942 |
1
|
1863
|
|
GO:0007476
|
imaginal disc-derived wing morphogenesis
|
0.816751 |
0.816751 |
2
|
254
|
|
GO:0002009
|
morphogenesis of an epithelium
|
0.825665 |
0.825665 |
2
|
289
|
|
GO:0001736
|
establishment of planar polarity
|
0.853933 |
0.853933 |
1
|
76
|
|
GO:0044446
|
intracellular organelle part
|
0.869797 |
0.869797 |
1
|
2162
|
|
GO:0043229
|
intracellular organelle
|
0.899971 |
0.899971 |
2
|
3530
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.900589 |
0.900589 |
1
|
620
|
|
GO:0045449
|
regulation of transcription
|
0.902327 |
0.902327 |
1
|
771
|
|
GO:0005737
|
cytoplasm
|
0.904274 |
0.904274 |
1
|
2378
|
|
GO:0002165
|
instar larval or pupal development
|
0.910028 |
0.910028 |
2
|
477
|
|
GO:0030154
|
cell differentiation
|
0.942825 |
0.942825 |
2
|
1239
|
|
GO:0035114
|
imaginal disc-derived appendage morphogenesis
|
0.951564 |
0.951564 |
2
|
279
|
|
GO:0035315
|
hair cell differentiation
|
0.956522 |
0.956522 |
1
|
44
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0003899
|
DNA-directed RNA polymerase activity
|
0.971429 |
0.971429 |
1
|
34
|
|
GO:0048737
|
imaginal disc-derived appendage development
|
0.972175 |
0.972175 |
2
|
282
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0035110
|
leg morphogenesis
|
0.978723 |
0.978723 |
1
|
46
|
|
GO:0007424
|
open tracheal system development
|
0.995984 |
0.995984 |
1
|
248
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0000428
|
DNA-directed RNA polymerase complex
|
1.00000 |
1.00000 |
1
|
28
|
|
GO:0003700
|
sequence-specific DNA binding transcription factor activity
|
1.00000 |
1.00000 |
1
|
395
|