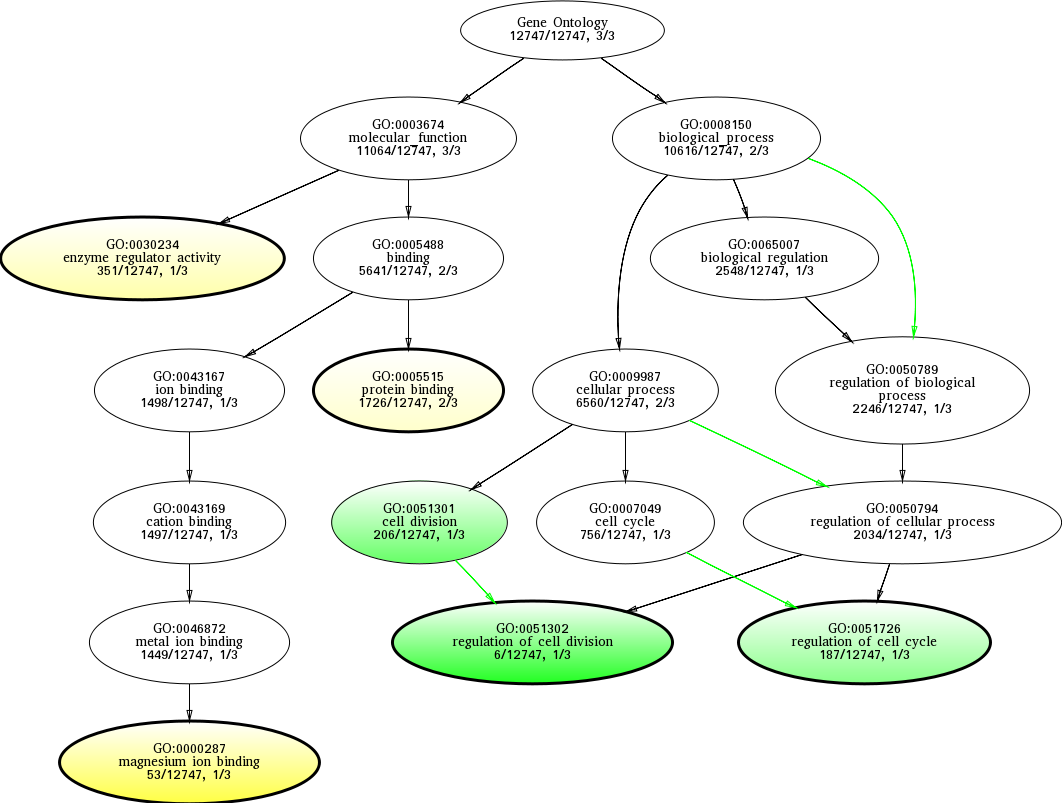
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0051302
|
regulation of cell division
|
0.00284360 |
0.00284360 |
1
|
6
|
|
GO:0000287
|
magnesium ion binding
|
0.0365769 |
0.0365769 |
1
|
53
|
|
GO:0051301
|
cell division
|
0.0618234 |
0.0618234 |
1
|
206
|
|
GO:0051726
|
regulation of cell cycle
|
0.0752212 |
0.0752212 |
1
|
187
|
|
GO:0030234
|
enzyme regulator activity
|
0.0921942 |
0.0921942 |
1
|
351
|
|
GO:0005515
|
protein binding
|
0.0935825 |
0.0935825 |
2
|
1726
|
|
GO:0016044
|
cellular membrane organization
|
0.102136 |
0.102136 |
1
|
344
|
|
GO:0016192
|
vesicle-mediated transport
|
0.121581 |
0.121581 |
1
|
430
|
|
GO:0006793
|
phosphorus metabolic process
|
0.145779 |
0.145779 |
1
|
556
|
|
GO:0019207
|
kinase regulator activity
|
0.162393 |
0.162393 |
1
|
57
|
|
GO:0061024
|
membrane organization
|
0.168129 |
0.168129 |
1
|
345
|
|
GO:0043412
|
macromolecule modification
|
0.201784 |
0.201784 |
1
|
724
|
|
GO:0007049
|
cell cycle
|
0.217222 |
0.217222 |
1
|
756
|
|
GO:0016740
|
transferase activity
|
0.250852 |
0.250852 |
1
|
1030
|
|
GO:0001882
|
nucleoside binding
|
0.258670 |
0.258670 |
1
|
784
|
|
GO:0051234
|
establishment of localization
|
0.270545 |
0.270545 |
1
|
1549
|
|
GO:0051179
|
localization
|
0.325313 |
0.325313 |
1
|
1896
|
|
GO:0016538
|
cyclin-dependent protein kinase regulator activity
|
0.327273 |
0.327273 |
1
|
18
|
|
GO:0006468
|
protein phosphorylation
|
0.335758 |
0.335758 |
1
|
277
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.357722 |
0.357722 |
1
|
2108
|
|
GO:0000166
|
nucleotide binding
|
0.374077 |
0.374077 |
1
|
1178
|
|
GO:0050789
|
regulation of biological process
|
0.378390 |
0.378390 |
1
|
2246
|
|
GO:0009987
|
cellular process
|
0.381822 |
0.381822 |
2
|
6560
|
|
GO:0065007
|
biological regulation
|
0.422440 |
0.422440 |
1
|
2548
|
|
GO:0044267
|
cellular protein metabolic process
|
0.440058 |
0.440058 |
1
|
1505
|
|
GO:0006464
|
protein modification process
|
0.443005 |
0.443005 |
1
|
684
|
|
GO:0043167
|
ion binding
|
0.460626 |
0.460626 |
1
|
1498
|
|
GO:0019538
|
protein metabolic process
|
0.475452 |
0.475452 |
1
|
2053
|
|
GO:0016772
|
transferase activity, transferring phosphorus-containing groups
|
0.480583 |
0.480583 |
1
|
495
|
|
GO:0005488
|
binding
|
0.514777 |
0.514777 |
2
|
5641
|
|
GO:0050794
|
regulation of cellular process
|
0.518911 |
0.518911 |
1
|
2034
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.639129 |
0.639129 |
1
|
2878
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0006897
|
endocytosis
|
0.655814 |
0.655814 |
1
|
282
|
|
GO:0006911
|
phagocytosis, engulfment
|
0.680851 |
0.680851 |
1
|
192
|
|
GO:0004674
|
protein serine/threonine kinase activity
|
0.686207 |
0.686207 |
1
|
199
|
|
GO:0043170
|
macromolecule metabolic process
|
0.690797 |
0.690797 |
1
|
3588
|
|
GO:0006909
|
phagocytosis
|
0.723404 |
0.723404 |
1
|
204
|
|
GO:0004672
|
protein kinase activity
|
0.728643 |
0.728643 |
1
|
290
|
|
GO:0016773
|
phosphotransferase activity, alcohol group as acceptor
|
0.735354 |
0.735354 |
1
|
364
|
|
GO:0008152
|
metabolic process
|
0.739170 |
0.739170 |
1
|
5194
|
|
GO:0044237
|
cellular metabolic process
|
0.740940 |
0.740940 |
1
|
3814
|
|
GO:0032553
|
ribonucleotide binding
|
0.746180 |
0.746180 |
1
|
879
|
|
GO:0003824
|
catalytic activity
|
0.751316 |
0.751316 |
1
|
4106
|
|
GO:0032559
|
adenyl ribonucleotide binding
|
0.757705 |
0.757705 |
1
|
713
|
|
GO:0016310
|
phosphorylation
|
0.758993 |
0.758993 |
1
|
422
|
|
GO:0016301
|
kinase activity
|
0.795960 |
0.795960 |
1
|
394
|
|
GO:0017076
|
purine nucleotide binding
|
0.803056 |
0.803056 |
1
|
946
|
|
GO:0030554
|
adenyl nucleotide binding
|
0.819239 |
0.819239 |
1
|
775
|
|
GO:0010324
|
membrane invagination
|
0.819767 |
0.819767 |
1
|
282
|
|
GO:0044238
|
primary metabolic process
|
0.819985 |
0.819985 |
1
|
4259
|
|
GO:0008150
|
biological_process
|
0.925522 |
0.925522 |
2
|
10616
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.929175 |
0.929175 |
1
|
879
|
|
GO:0019887
|
protein kinase regulator activity
|
0.964912 |
0.964912 |
1
|
55
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0001883
|
purine nucleoside binding
|
0.992347 |
0.992347 |
1
|
778
|
|
GO:0005524
|
ATP binding
|
0.998597 |
0.998597 |
1
|
712
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0006796
|
phosphate metabolic process
|
1.00000 |
1.00000 |
1
|
556
|