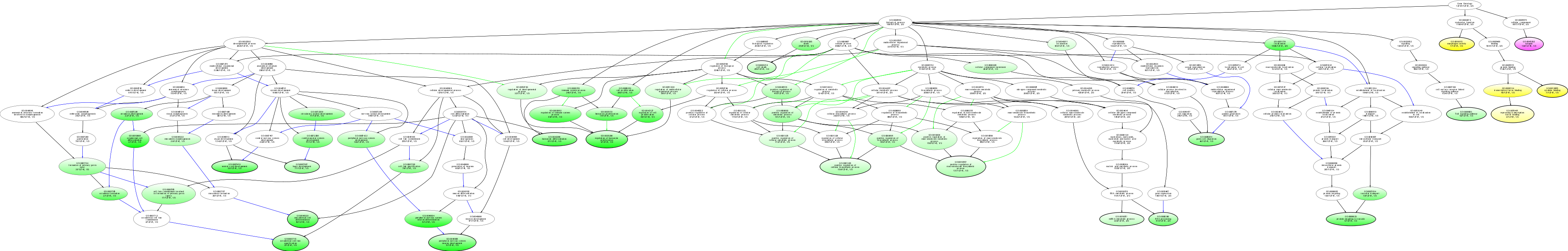
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0010668
|
ectodermal cell differentiation
|
0.00161031 |
0.00161031 |
1
|
2
|
|
GO:0016015
|
morphogen activity
|
0.00189702 |
0.00189702 |
1
|
7
|
|
GO:0019955
|
cytokine binding
|
0.00405562 |
0.00405562 |
1
|
7
|
|
GO:0035206
|
regulation of hemocyte proliferation
|
0.00817996 |
0.00817996 |
1
|
4
|
|
GO:0048934
|
peripheral nervous system neuron differentiation
|
0.00834725 |
0.00834725 |
1
|
5
|
|
GO:0048935
|
peripheral nervous system neuron development
|
0.0106157 |
0.0106157 |
1
|
5
|
|
GO:0048333
|
mesodermal cell differentiation
|
0.0128617 |
0.0128617 |
1
|
16
|
|
GO:0001715
|
ectodermal cell fate specification
|
0.0253165 |
0.0253165 |
1
|
2
|
|
GO:0007419
|
ventral cord development
|
0.0257410 |
0.0257410 |
1
|
33
|
|
GO:0008134
|
transcription factor binding
|
0.0260718 |
0.0260718 |
1
|
45
|
|
GO:0042386
|
hemocyte differentiation
|
0.0264853 |
0.0264853 |
1
|
37
|
|
GO:0002682
|
regulation of immune system process
|
0.0294494 |
0.0294494 |
1
|
69
|
|
GO:0042127
|
regulation of cell proliferation
|
0.0311005 |
0.0311005 |
1
|
65
|
|
GO:0051179
|
localization
|
0.0318835 |
0.0318835 |
2
|
1896
|
|
GO:0006623
|
protein targeting to vacuole
|
0.0335570 |
0.0335570 |
1
|
5
|
|
GO:0035172
|
hemocyte proliferation
|
0.0337349 |
0.0337349 |
1
|
14
|
|
GO:0008283
|
cell proliferation
|
0.0373258 |
0.0373258 |
1
|
200
|
|
GO:0007398
|
ectoderm development
|
0.0394973 |
0.0394973 |
1
|
22
|
|
GO:0001705
|
ectoderm formation
|
0.0400000 |
0.0400000 |
1
|
2
|
|
GO:0045202
|
synapse
|
0.0449093 |
0.0449093 |
1
|
134
|
|
GO:0072359
|
circulatory system development
|
0.0454009 |
0.0454009 |
1
|
77
|
|
GO:0072358
|
cardiovascular system development
|
0.0454009 |
0.0454009 |
1
|
77
|
|
GO:0002376
|
immune system process
|
0.0456263 |
0.0456263 |
1
|
245
|
|
GO:0006396
|
RNA processing
|
0.0458312 |
0.0458312 |
2
|
414
|
|
GO:0008354
|
germ cell migration
|
0.0472279 |
0.0472279 |
1
|
46
|
|
GO:0016265
|
death
|
0.0482013 |
0.0482013 |
1
|
259
|
|
GO:0007034
|
vacuolar transport
|
0.0539773 |
0.0539773 |
1
|
19
|
|
GO:0007422
|
peripheral nervous system development
|
0.0589623 |
0.0589623 |
1
|
100
|
|
GO:0001708
|
cell fate specification
|
0.0619122 |
0.0619122 |
1
|
79
|
|
GO:0007507
|
heart development
|
0.0619968 |
0.0619968 |
1
|
77
|
|
GO:0001704
|
formation of primary germ layer
|
0.0633947 |
0.0633947 |
1
|
31
|
|
GO:0060795
|
cell fate commitment involved in formation of primary germ layers
|
0.0636704 |
0.0636704 |
1
|
17
|
|
GO:0070491
|
repressing transcription factor binding
|
0.0666667 |
0.0666667 |
1
|
3
|
|
GO:0009893
|
positive regulation of metabolic process
|
0.0671169 |
0.0671169 |
1
|
181
|
|
GO:0008063
|
Toll signaling pathway
|
0.0673077 |
0.0673077 |
1
|
42
|
|
GO:0048518
|
positive regulation of biological process
|
0.0757538 |
0.0757538 |
1
|
410
|
|
GO:0009891
|
positive regulation of biosynthetic process
|
0.0760818 |
0.0760818 |
1
|
160
|
|
GO:0008219
|
cell death
|
0.0762386 |
0.0762386 |
1
|
255
|
|
GO:0031328
|
positive regulation of cellular biosynthetic process
|
0.0777886 |
0.0777886 |
1
|
159
|
|
GO:0010557
|
positive regulation of macromolecule biosynthetic process
|
0.0789632 |
0.0789632 |
1
|
131
|
|
GO:0048332
|
mesoderm morphogenesis
|
0.0794521 |
0.0794521 |
1
|
29
|
|
GO:0006928
|
cellular component movement
|
0.0803367 |
0.0803367 |
1
|
269
|
|
GO:0016071
|
mRNA metabolic process
|
0.0808458 |
0.0808458 |
2
|
339
|
|
GO:0010604
|
positive regulation of macromolecule metabolic process
|
0.0814533 |
0.0814533 |
1
|
150
|
|
GO:0051239
|
regulation of multicellular organismal process
|
0.0844424 |
0.0844424 |
1
|
368
|
|
GO:0050793
|
regulation of developmental process
|
0.0844959 |
0.0844959 |
1
|
321
|
|
GO:0031325
|
positive regulation of cellular metabolic process
|
0.0858406 |
0.0858406 |
1
|
172
|
|
GO:0040011
|
locomotion
|
0.0891086 |
0.0891086 |
1
|
484
|
|
GO:0001071
|
nucleic acid binding transcription factor activity
|
0.103335 |
0.103335 |
1
|
395
|
|
GO:0050684
|
regulation of mRNA processing
|
0.105613 |
0.105613 |
1
|
65
|
|
GO:0051173
|
positive regulation of nitrogen compound metabolic process
|
0.108443 |
0.108443 |
1
|
119
|
|
GO:0048522
|
positive regulation of cellular process
|
0.110028 |
0.110028 |
1
|
372
|
|
GO:0043484
|
regulation of RNA splicing
|
0.110325 |
0.110325 |
1
|
66
|
|
GO:0030528
|
transcription regulator activity
|
0.112632 |
0.112632 |
1
|
432
|
|
GO:0006955
|
immune response
|
0.113493 |
0.113493 |
1
|
180
|
|
GO:0045893
|
positive regulation of transcription, DNA-dependent
|
0.114685 |
0.114685 |
1
|
82
|
|
GO:0001712
|
ectodermal cell fate commitment
|
0.117647 |
0.117647 |
1
|
2
|
|
GO:0010628
|
positive regulation of gene expression
|
0.123263 |
0.123263 |
1
|
122
|
|
GO:0045935
|
positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.125793 |
0.125793 |
1
|
119
|
|
GO:0045941
|
positive regulation of transcription
|
0.127636 |
0.127636 |
1
|
115
|
|
GO:0048519
|
negative regulation of biological process
|
0.128590 |
0.128590 |
1
|
706
|
|
GO:0009890
|
negative regulation of biosynthetic process
|
0.129715 |
0.129715 |
1
|
282
|
|
GO:0051254
|
positive regulation of RNA metabolic process
|
0.130868 |
0.130868 |
1
|
83
|
|
GO:0007501
|
mesodermal cell fate specification
|
0.130952 |
0.130952 |
1
|
11
|
|
GO:0005488
|
binding
|
0.132501 |
0.132501 |
3
|
5641
|
|
GO:0009892
|
negative regulation of metabolic process
|
0.133617 |
0.133617 |
1
|
372
|
|
GO:0042387
|
plasmatocyte differentiation
|
0.135135 |
0.135135 |
1
|
5
|
|
GO:0007417
|
central nervous system development
|
0.135613 |
0.135613 |
1
|
230
|
|
GO:0031327
|
negative regulation of cellular biosynthetic process
|
0.136166 |
0.136166 |
1
|
282
|
|
GO:0048598
|
embryonic morphogenesis
|
0.136979 |
0.136979 |
1
|
243
|
|
GO:0016563
|
transcription activator activity
|
0.141204 |
0.141204 |
1
|
61
|
|
GO:0070379
|
high mobility group box 1 binding
|
0.142857 |
0.142857 |
1
|
1
|
|
GO:0000122
|
negative regulation of transcription from RNA polymerase II promoter
|
0.145455 |
0.145455 |
1
|
56
|
|
GO:0031324
|
negative regulation of cellular metabolic process
|
0.152355 |
0.152355 |
1
|
319
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.162334 |
0.162334 |
2
|
2093
|
|
GO:0010558
|
negative regulation of macromolecule biosynthetic process
|
0.165094 |
0.165094 |
1
|
280
|
|
GO:0005681
|
spliceosomal complex
|
0.167598 |
0.167598 |
1
|
180
|
|
GO:0045184
|
establishment of protein localization
|
0.168623 |
0.168623 |
1
|
273
|
|
GO:0051641
|
cellular localization
|
0.169056 |
0.169056 |
1
|
607
|
|
GO:0045944
|
positive regulation of transcription from RNA polymerase II promoter
|
0.170213 |
0.170213 |
1
|
48
|
|
GO:0007498
|
mesoderm development
|
0.170557 |
0.170557 |
1
|
95
|
|
GO:0016070
|
RNA metabolic process
|
0.171170 |
0.171170 |
2
|
1191
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.172797 |
0.172797 |
2
|
1789
|
|
GO:2000113
|
negative regulation of cellular macromolecule biosynthetic process
|
0.172946 |
0.172946 |
1
|
280
|
|
GO:0048610
|
cellular process involved in reproduction
|
0.173346 |
0.173346 |
1
|
613
|
|
GO:0048646
|
anatomical structure formation involved in morphogenesis
|
0.176649 |
0.176649 |
1
|
466
|
|
GO:0015031
|
protein transport
|
0.177333 |
0.177333 |
1
|
266
|
|
GO:0000003
|
reproduction
|
0.183280 |
0.183280 |
1
|
1022
|
|
GO:0022414
|
reproductive process
|
0.183280 |
0.183280 |
1
|
1022
|
|
GO:0048523
|
negative regulation of cellular process
|
0.183975 |
0.183975 |
1
|
635
|
|
GO:0031594
|
neuromuscular junction
|
0.186567 |
0.186567 |
1
|
25
|
|
GO:0010605
|
negative regulation of macromolecule metabolic process
|
0.187925 |
0.187925 |
1
|
356
|
|
GO:0048729
|
tissue morphogenesis
|
0.194151 |
0.194151 |
1
|
312
|
|
GO:0045165
|
cell fate commitment
|
0.199843 |
0.199843 |
1
|
255
|
|
GO:0007389
|
pattern specification process
|
0.203563 |
0.203563 |
1
|
537
|
|
GO:0030529
|
ribonucleoprotein complex
|
0.210589 |
0.210589 |
1
|
510
|
|
GO:0016564
|
transcription repressor activity
|
0.215278 |
0.215278 |
1
|
93
|
|
GO:0051172
|
negative regulation of nitrogen compound metabolic process
|
0.216720 |
0.216720 |
1
|
250
|
|
GO:0046907
|
intracellular transport
|
0.227832 |
0.227832 |
1
|
352
|
|
GO:0008152
|
metabolic process
|
0.239353 |
0.239353 |
2
|
5194
|
|
GO:0023052
|
signaling
|
0.240473 |
0.240473 |
1
|
1364
|
|
GO:0044237
|
cellular metabolic process
|
0.241038 |
0.241038 |
2
|
3814
|
|
GO:0009790
|
embryo development
|
0.242987 |
0.242987 |
1
|
641
|
|
GO:0016481
|
negative regulation of transcription
|
0.243724 |
0.243724 |
1
|
233
|
|
GO:0009888
|
tissue development
|
0.245916 |
0.245916 |
1
|
557
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.248286 |
0.248286 |
2
|
1959
|
|
GO:0006622
|
protein targeting to lysosome
|
0.250000 |
0.250000 |
1
|
4
|
|
GO:0045934
|
negative regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.252452 |
0.252452 |
1
|
250
|
|
GO:0003676
|
nucleic acid binding
|
0.253252 |
0.253252 |
2
|
1855
|
|
GO:0051674
|
localization of cell
|
0.258248 |
0.258248 |
1
|
263
|
|
GO:0045892
|
negative regulation of transcription, DNA-dependent
|
0.266030 |
0.266030 |
1
|
195
|
|
GO:0050896
|
response to stimulus
|
0.270384 |
0.270384 |
1
|
1548
|
|
GO:0051234
|
establishment of localization
|
0.270545 |
0.270545 |
1
|
1549
|
|
GO:0009950
|
dorsal/ventral axis specification
|
0.274306 |
0.274306 |
1
|
79
|
|
GO:0010629
|
negative regulation of gene expression
|
0.276112 |
0.276112 |
1
|
288
|
|
GO:0048609
|
multicellular organismal reproductive process
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0032504
|
multicellular organism reproduction
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0010467
|
gene expression
|
0.282416 |
0.282416 |
2
|
1907
|
|
GO:0006357
|
regulation of transcription from RNA polymerase II promoter
|
0.282934 |
0.282934 |
1
|
189
|
|
GO:0009953
|
dorsal/ventral pattern formation
|
0.286561 |
0.286561 |
1
|
145
|
|
GO:0051253
|
negative regulation of RNA metabolic process
|
0.287333 |
0.287333 |
1
|
202
|
|
GO:0009880
|
embryonic pattern specification
|
0.294591 |
0.294591 |
1
|
256
|
|
GO:0043565
|
sequence-specific DNA binding
|
0.300971 |
0.300971 |
1
|
248
|
|
GO:0051649
|
establishment of localization in cell
|
0.303951 |
0.303951 |
1
|
500
|
|
GO:0007369
|
gastrulation
|
0.312757 |
0.312757 |
1
|
76
|
|
GO:0006952
|
defense response
|
0.314050 |
0.314050 |
1
|
190
|
|
GO:0048869
|
cellular developmental process
|
0.322329 |
0.322329 |
1
|
1276
|
|
GO:0005575
|
cellular_component
|
0.330897 |
0.330897 |
3
|
8817
|
|
GO:0007351
|
tripartite regional subdivision
|
0.332016 |
0.332016 |
1
|
168
|
|
GO:0005623
|
cell
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0044464
|
cell part
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0043226
|
organelle
|
0.353651 |
0.353651 |
2
|
3537
|
|
GO:0006366
|
transcription from RNA polymerase II promoter
|
0.360913 |
0.360913 |
1
|
253
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.366791 |
0.366791 |
1
|
888
|
|
GO:0019222
|
regulation of metabolic process
|
0.376562 |
0.376562 |
1
|
1242
|
|
GO:0050789
|
regulation of biological process
|
0.378390 |
0.378390 |
1
|
2246
|
|
GO:0009987
|
cellular process
|
0.381822 |
0.381822 |
2
|
6560
|
|
GO:0000380
|
alternative nuclear mRNA splicing, via spliceosome
|
0.389190 |
0.389190 |
1
|
58
|
|
GO:0009952
|
anterior/posterior pattern formation
|
0.389328 |
0.389328 |
1
|
197
|
|
GO:0006950
|
response to stress
|
0.390827 |
0.390827 |
1
|
605
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.396252 |
0.396252 |
1
|
888
|
|
GO:0070727
|
cellular macromolecule localization
|
0.408300 |
0.408300 |
1
|
305
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.408435 |
0.408435 |
2
|
2878
|
|
GO:0005634
|
nucleus
|
0.408695 |
0.408695 |
2
|
1826
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.418200 |
0.418200 |
1
|
1039
|
|
GO:0048024
|
regulation of nuclear mRNA splicing, via spliceosome
|
0.419908 |
0.419908 |
1
|
64
|
|
GO:0065007
|
biological regulation
|
0.422440 |
0.422440 |
1
|
2548
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.429576 |
0.429576 |
1
|
1110
|
|
GO:0009798
|
axis specification
|
0.432030 |
0.432030 |
1
|
232
|
|
GO:0048666
|
neuron development
|
0.433702 |
0.433702 |
1
|
471
|
|
GO:0032502
|
developmental process
|
0.435255 |
0.435255 |
1
|
2638
|
|
GO:0030182
|
neuron differentiation
|
0.442292 |
0.442292 |
1
|
548
|
|
GO:0005622
|
intracellular
|
0.446165 |
0.446165 |
3
|
4734
|
|
GO:0008380
|
RNA splicing
|
0.447135 |
0.447135 |
2
|
277
|
|
GO:0022008
|
neurogenesis
|
0.452398 |
0.452398 |
1
|
632
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.456989 |
0.456989 |
1
|
850
|
|
GO:0048468
|
cell development
|
0.457419 |
0.457419 |
1
|
1042
|
|
GO:0044428
|
nuclear part
|
0.469235 |
0.469235 |
1
|
830
|
|
GO:0033036
|
macromolecule localization
|
0.476467 |
0.476467 |
1
|
524
|
|
GO:0043170
|
macromolecule metabolic process
|
0.477159 |
0.477159 |
2
|
3588
|
|
GO:0034613
|
cellular protein localization
|
0.478936 |
0.478936 |
1
|
216
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.486014 |
0.486014 |
1
|
1056
|
|
GO:0032774
|
RNA biosynthetic process
|
0.496008 |
0.496008 |
1
|
703
|
|
GO:0001703
|
gastrulation with mouth forming first
|
0.500000 |
0.500000 |
1
|
38
|
|
GO:0007399
|
nervous system development
|
0.500000 |
0.500000 |
1
|
848
|
|
GO:0000166
|
nucleotide binding
|
0.504833 |
0.504833 |
1
|
1178
|
|
GO:0006886
|
intracellular protein transport
|
0.504878 |
0.504878 |
1
|
207
|
|
GO:0048870
|
cell motility
|
0.512097 |
0.512097 |
1
|
254
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.513595 |
0.513595 |
1
|
850
|
|
GO:0050794
|
regulation of cellular process
|
0.518911 |
0.518911 |
1
|
2034
|
|
GO:0032501
|
multicellular organismal process
|
0.527040 |
0.527040 |
1
|
3315
|
|
GO:0006397
|
mRNA processing
|
0.538908 |
0.538908 |
2
|
318
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0000578
|
embryonic axis specification
|
0.552716 |
0.552716 |
1
|
173
|
|
GO:0032991
|
macromolecular complex
|
0.567899 |
0.567899 |
1
|
2151
|
|
GO:0044422
|
organelle part
|
0.572743 |
0.572743 |
1
|
2176
|
|
GO:0035282
|
segmentation
|
0.581028 |
0.581028 |
1
|
294
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0043167
|
ion binding
|
0.603911 |
0.603911 |
1
|
1498
|
|
GO:0008358
|
maternal determination of anterior/posterior axis, embryo
|
0.613095 |
0.613095 |
1
|
103
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.617799 |
0.617799 |
1
|
905
|
|
GO:0003702
|
RNA polymerase II transcription factor activity
|
0.618056 |
0.618056 |
1
|
267
|
|
GO:0006350
|
transcription
|
0.625331 |
0.625331 |
1
|
859
|
|
GO:0006915
|
apoptosis
|
0.633333 |
0.633333 |
1
|
152
|
|
GO:0009058
|
biosynthetic process
|
0.642905 |
0.642905 |
1
|
2090
|
|
GO:0043229
|
intracellular organelle
|
0.646541 |
0.646541 |
2
|
3530
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.646807 |
0.646807 |
1
|
1623
|
|
GO:0007166
|
cell surface receptor linked signaling pathway
|
0.652720 |
0.652720 |
1
|
624
|
|
GO:0043227
|
membrane-bounded organelle
|
0.653325 |
0.653325 |
2
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.653801 |
0.653801 |
2
|
2856
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0006605
|
protein targeting
|
0.657005 |
0.657005 |
1
|
136
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0005515
|
protein binding
|
0.665786 |
0.665786 |
1
|
1726
|
|
GO:0044238
|
primary metabolic process
|
0.672346 |
0.672346 |
2
|
4259
|
|
GO:0009948
|
anterior/posterior axis specification
|
0.675781 |
0.675781 |
1
|
173
|
|
GO:0071013
|
catalytic step 2 spliceosome
|
0.683333 |
0.683333 |
1
|
123
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.684327 |
0.684327 |
1
|
620
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.686903 |
0.686903 |
1
|
686
|
|
GO:0045449
|
regulation of transcription
|
0.687166 |
0.687166 |
1
|
771
|
|
GO:0003677
|
DNA binding
|
0.691225 |
0.691225 |
1
|
824
|
|
GO:0000398
|
nuclear mRNA splicing, via spliceosome
|
0.699264 |
0.699264 |
2
|
266
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.699519 |
0.699519 |
1
|
903
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0008104
|
protein localization
|
0.727099 |
0.727099 |
1
|
381
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.736132 |
0.736132 |
2
|
1535
|
|
GO:0010468
|
regulation of gene expression
|
0.739954 |
0.739954 |
1
|
973
|
|
GO:0044446
|
intracellular organelle part
|
0.743053 |
0.743053 |
1
|
2162
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.746994 |
0.746994 |
1
|
1617
|
|
GO:0044249
|
cellular biosynthetic process
|
0.776153 |
0.776153 |
1
|
2034
|
|
GO:0007350
|
blastoderm segmentation
|
0.779221 |
0.779221 |
1
|
240
|
|
GO:0008270
|
zinc ion binding
|
0.781440 |
0.781440 |
1
|
901
|
|
GO:0007041
|
lysosomal transport
|
0.789474 |
0.789474 |
1
|
15
|
|
GO:0005737
|
cytoplasm
|
0.790684 |
0.790684 |
1
|
2378
|
|
GO:0044424
|
intracellular part
|
0.793631 |
0.793631 |
2
|
4384
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.814170 |
0.814170 |
1
|
701
|
|
GO:0071011
|
precatalytic spliceosome
|
0.822222 |
0.822222 |
1
|
148
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0019953
|
sexual reproduction
|
0.859100 |
0.859100 |
1
|
878
|
|
GO:0001707
|
mesoderm formation
|
0.875000 |
0.875000 |
1
|
28
|
|
GO:0007276
|
gamete generation
|
0.883697 |
0.883697 |
1
|
851
|
|
GO:0000381
|
regulation of alternative nuclear mRNA splicing, via spliceosome
|
0.890625 |
0.890625 |
1
|
57
|
|
GO:0008150
|
biological_process
|
0.925522 |
0.925522 |
2
|
10616
|
|
GO:0000375
|
RNA splicing, via transesterification reactions
|
0.928975 |
0.928975 |
2
|
267
|
|
GO:0012501
|
programmed cell death
|
0.941176 |
0.941176 |
1
|
240
|
|
GO:0001710
|
mesodermal cell fate commitment
|
0.941176 |
0.941176 |
1
|
16
|
|
GO:0003002
|
regionalization
|
0.942272 |
0.942272 |
1
|
506
|
|
GO:0008595
|
anterior/posterior axis specification, embryo
|
0.943820 |
0.943820 |
1
|
168
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0016477
|
cell migration
|
0.964567 |
0.964567 |
1
|
245
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0000377
|
RNA splicing, via transesterification reactions with bulged adenosine as nucleophile
|
0.992509 |
0.992509 |
2
|
266
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0003700
|
sequence-specific DNA binding transcription factor activity
|
1.00000 |
1.00000 |
1
|
395
|
|
GO:0010004
|
gastrulation involving germ band extension
|
1.00000 |
1.00000 |
1
|
38
|