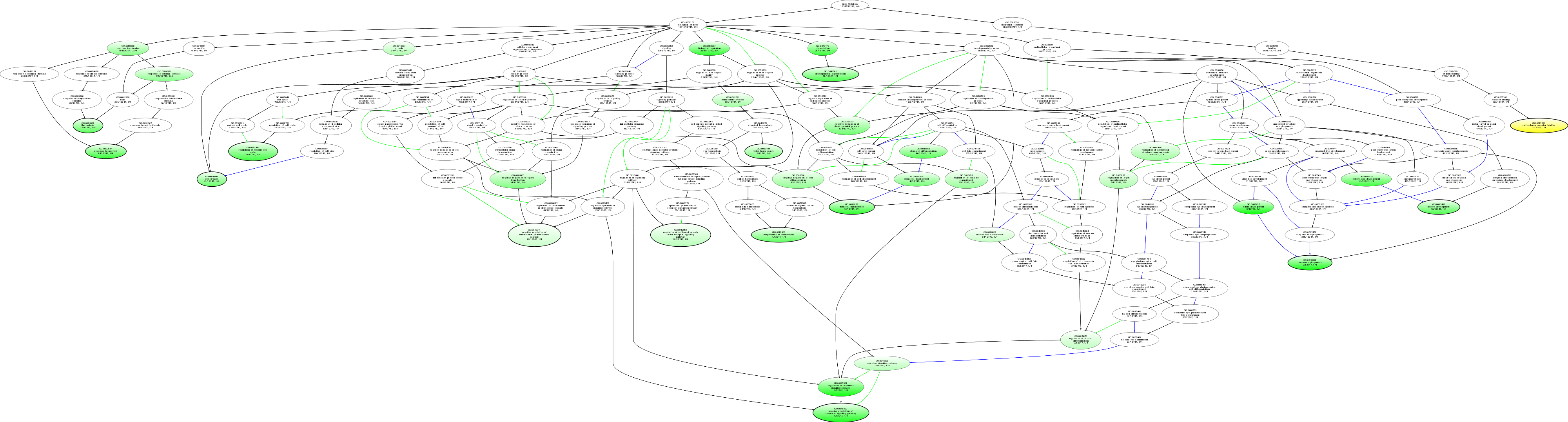
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0007477
|
notum development
|
0.00220751 |
0.00220751 |
1
|
5
|
|
GO:0045501
|
regulation of sevenless signaling pathway
|
0.00284900 |
0.00284900 |
1
|
1
|
|
GO:0048802
|
notum morphogenesis
|
0.00484262 |
0.00484262 |
1
|
2
|
|
GO:0017098
|
sulfonylurea receptor binding
|
0.00578035 |
0.00578035 |
1
|
1
|
|
GO:0045873
|
negative regulation of sevenless signaling pathway
|
0.00787402 |
0.00787402 |
1
|
1
|
|
GO:0065007
|
biological regulation
|
0.0138142 |
0.0138142 |
3
|
2548
|
|
GO:0019827
|
stem cell maintenance
|
0.0174375 |
0.0174375 |
1
|
46
|
|
GO:0043473
|
pigmentation
|
0.0213256 |
0.0213256 |
1
|
76
|
|
GO:0007584
|
response to nutrient
|
0.0217054 |
0.0217054 |
1
|
14
|
|
GO:0035216
|
haltere disc development
|
0.0235546 |
0.0235546 |
1
|
11
|
|
GO:0051445
|
regulation of meiotic cell cycle
|
0.0237530 |
0.0237530 |
1
|
10
|
|
GO:0048066
|
developmental pigmentation
|
0.0268939 |
0.0268939 |
1
|
71
|
|
GO:0007482
|
haltere development
|
0.0354610 |
0.0354610 |
1
|
10
|
|
GO:0048863
|
stem cell differentiation
|
0.0371267 |
0.0371267 |
1
|
46
|
|
GO:0043052
|
thermotaxis
|
0.0389610 |
0.0389610 |
1
|
12
|
|
GO:0010960
|
magnesium ion homeostasis
|
0.0400000 |
0.0400000 |
1
|
1
|
|
GO:0016049
|
cell growth
|
0.0409182 |
0.0409182 |
1
|
91
|
|
GO:0051093
|
negative regulation of developmental process
|
0.0423208 |
0.0423208 |
1
|
124
|
|
GO:0048864
|
stem cell development
|
0.0441459 |
0.0441459 |
1
|
46
|
|
GO:0030104
|
water homeostasis
|
0.0503083 |
0.0503083 |
1
|
2
|
|
GO:0009968
|
negative regulation of signal transduction
|
0.0547504 |
0.0547504 |
1
|
34
|
|
GO:0050896
|
response to stimulus
|
0.0575624 |
0.0575624 |
2
|
1548
|
|
GO:0009605
|
response to external stimulus
|
0.0585655 |
0.0585655 |
2
|
375
|
|
GO:0045596
|
negative regulation of cell differentiation
|
0.0604154 |
0.0604154 |
1
|
96
|
|
GO:0010453
|
regulation of cell fate commitment
|
0.0612745 |
0.0612745 |
1
|
25
|
|
GO:0022603
|
regulation of anatomical structure morphogenesis
|
0.0618812 |
0.0618812 |
1
|
100
|
|
GO:0042592
|
homeostatic process
|
0.0655384 |
0.0655384 |
2
|
187
|
|
GO:2000027
|
regulation of organ morphogenesis
|
0.0659341 |
0.0659341 |
1
|
54
|
|
GO:0040007
|
growth
|
0.0681951 |
0.0681951 |
1
|
247
|
|
GO:0045500
|
sevenless signaling pathway
|
0.0694444 |
0.0694444 |
1
|
10
|
|
GO:0042058
|
regulation of epidermal growth factor receptor signaling pathway
|
0.0802292 |
0.0802292 |
1
|
28
|
|
GO:0045676
|
regulation of R7 cell differentiation
|
0.0813953 |
0.0813953 |
1
|
7
|
|
GO:0048663
|
neuron fate commitment
|
0.0942563 |
0.0942563 |
1
|
64
|
|
GO:0010741
|
negative regulation of intracellular protein kinase cascade
|
0.0984456 |
0.0984456 |
1
|
19
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.102602 |
0.102602 |
2
|
2108
|
|
GO:0042059
|
negative regulation of epidermal growth factor receptor signaling pathway
|
0.109677 |
0.109677 |
1
|
17
|
|
GO:2000026
|
regulation of multicellular organismal development
|
0.109792 |
0.109792 |
1
|
259
|
|
GO:0048736
|
appendage development
|
0.110896 |
0.110896 |
1
|
286
|
|
GO:0008586
|
imaginal disc-derived wing vein morphogenesis
|
0.114078 |
0.114078 |
1
|
47
|
|
GO:0050789
|
regulation of biological process
|
0.115315 |
0.115315 |
2
|
2246
|
|
GO:0007005
|
mitochondrion organization
|
0.118165 |
0.118165 |
1
|
72
|
|
GO:0046532
|
regulation of photoreceptor cell differentiation
|
0.118812 |
0.118812 |
1
|
24
|
|
GO:0032535
|
regulation of cellular component size
|
0.122375 |
0.122375 |
1
|
130
|
|
GO:0035467
|
negative regulation of signaling pathway
|
0.122789 |
0.122789 |
1
|
118
|
|
GO:0046426
|
negative regulation of JAK-STAT cascade
|
0.125000 |
0.125000 |
1
|
4
|
|
GO:0023014
|
signal transmission via phosphorylation event
|
0.126316 |
0.126316 |
1
|
96
|
|
GO:0040011
|
locomotion
|
0.130645 |
0.130645 |
1
|
484
|
|
GO:0050767
|
regulation of neurogenesis
|
0.131004 |
0.131004 |
1
|
90
|
|
GO:0045664
|
regulation of neuron differentiation
|
0.131907 |
0.131907 |
1
|
74
|
|
GO:0010627
|
regulation of intracellular protein kinase cascade
|
0.132075 |
0.132075 |
1
|
49
|
|
GO:0051960
|
regulation of nervous system development
|
0.132196 |
0.132196 |
1
|
124
|
|
GO:0010648
|
negative regulation of cell communication
|
0.136511 |
0.136511 |
1
|
126
|
|
GO:0023051
|
regulation of signaling process
|
0.139523 |
0.139523 |
1
|
171
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.142259 |
0.142259 |
2
|
1562
|
|
GO:0055069
|
zinc ion homeostasis
|
0.142857 |
0.142857 |
1
|
2
|
|
GO:0051726
|
regulation of cell cycle
|
0.144812 |
0.144812 |
1
|
187
|
|
GO:0072511
|
divalent inorganic cation transport
|
0.149798 |
0.149798 |
1
|
37
|
|
GO:0046425
|
regulation of JAK-STAT cascade
|
0.157895 |
0.157895 |
1
|
9
|
|
GO:0060284
|
regulation of cell development
|
0.159091 |
0.159091 |
1
|
168
|
|
GO:0051239
|
regulation of multicellular organismal process
|
0.161772 |
0.161772 |
1
|
368
|
|
GO:0050793
|
regulation of developmental process
|
0.161873 |
0.161873 |
1
|
321
|
|
GO:0045595
|
regulation of cell differentiation
|
0.167859 |
0.167859 |
1
|
231
|
|
GO:0007259
|
JAK-STAT cascade
|
0.177083 |
0.177083 |
1
|
17
|
|
GO:0048878
|
chemical homeostasis
|
0.177161 |
0.177161 |
2
|
79
|
|
GO:0009991
|
response to extracellular stimulus
|
0.183016 |
0.183016 |
1
|
36
|
|
GO:0035107
|
appendage morphogenesis
|
0.184365 |
0.184365 |
1
|
283
|
|
GO:0048519
|
negative regulation of biological process
|
0.186553 |
0.186553 |
1
|
706
|
|
GO:0010646
|
regulation of cell communication
|
0.186653 |
0.186653 |
1
|
214
|
|
GO:0009791
|
post-embryonic development
|
0.189538 |
0.189538 |
1
|
500
|
|
GO:0005102
|
receptor binding
|
0.190469 |
0.190469 |
1
|
173
|
|
GO:0007017
|
microtubule-based process
|
0.191232 |
0.191232 |
1
|
448
|
|
GO:0055085
|
transmembrane transport
|
0.191633 |
0.191633 |
1
|
469
|
|
GO:0007154
|
cell communication
|
0.196383 |
0.196383 |
1
|
461
|
|
GO:0065008
|
regulation of biological quality
|
0.198629 |
0.198629 |
2
|
729
|
|
GO:0023057
|
negative regulation of signaling process
|
0.198830 |
0.198830 |
1
|
34
|
|
GO:0023046
|
signaling process
|
0.199779 |
0.199779 |
1
|
760
|
|
GO:0045165
|
cell fate commitment
|
0.199843 |
0.199843 |
1
|
255
|
|
GO:0006811
|
ion transport
|
0.221109 |
0.221109 |
1
|
331
|
|
GO:0005515
|
protein binding
|
0.223526 |
0.223526 |
2
|
1726
|
|
GO:0050794
|
regulation of cellular process
|
0.224039 |
0.224039 |
2
|
2034
|
|
GO:0070838
|
divalent metal ion transport
|
0.227848 |
0.227848 |
1
|
36
|
|
GO:0032501
|
multicellular organismal process
|
0.231607 |
0.231607 |
2
|
3315
|
|
GO:0009987
|
cellular process
|
0.235914 |
0.235914 |
3
|
6560
|
|
GO:0035466
|
regulation of signaling pathway
|
0.237323 |
0.237323 |
1
|
324
|
|
GO:0016740
|
transferase activity
|
0.250852 |
0.250852 |
1
|
1030
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.259446 |
0.259446 |
1
|
412
|
|
GO:0000003
|
reproduction
|
0.261920 |
0.261920 |
1
|
1022
|
|
GO:0022414
|
reproductive process
|
0.261920 |
0.261920 |
1
|
1022
|
|
GO:0048523
|
negative regulation of cellular process
|
0.262869 |
0.262869 |
1
|
635
|
|
GO:0046873
|
metal ion transmembrane transporter activity
|
0.268293 |
0.268293 |
1
|
110
|
|
GO:0022402
|
cell cycle process
|
0.270667 |
0.270667 |
1
|
655
|
|
GO:0007552
|
metamorphosis
|
0.273794 |
0.273794 |
1
|
420
|
|
GO:0005215
|
transporter activity
|
0.274270 |
0.274270 |
1
|
852
|
|
GO:0048569
|
post-embryonic organ development
|
0.276167 |
0.276167 |
1
|
343
|
|
GO:0022803
|
passive transmembrane transporter activity
|
0.276860 |
0.276860 |
1
|
201
|
|
GO:0007052
|
mitotic spindle organization
|
0.285513 |
0.285513 |
1
|
203
|
|
GO:0046530
|
photoreceptor cell differentiation
|
0.295620 |
0.295620 |
1
|
162
|
|
GO:0009628
|
response to abiotic stimulus
|
0.302411 |
0.302411 |
1
|
255
|
|
GO:0007243
|
intracellular protein kinase cascade
|
0.304762 |
0.304762 |
1
|
96
|
|
GO:0022838
|
substrate-specific channel activity
|
0.307448 |
0.307448 |
1
|
194
|
|
GO:0007049
|
cell cycle
|
0.307460 |
0.307460 |
1
|
756
|
|
GO:0009966
|
regulation of signal transduction
|
0.312044 |
0.312044 |
1
|
171
|
|
GO:0045466
|
R7 cell differentiation
|
0.313433 |
0.313433 |
1
|
42
|
|
GO:0007167
|
enzyme linked receptor protein signaling pathway
|
0.315705 |
0.315705 |
1
|
197
|
|
GO:0007465
|
R7 cell fate commitment
|
0.323529 |
0.323529 |
1
|
22
|
|
GO:0005488
|
binding
|
0.327389 |
0.327389 |
3
|
5641
|
|
GO:0009266
|
response to temperature stimulus
|
0.333333 |
0.333333 |
1
|
85
|
|
GO:0023052
|
signaling
|
0.338079 |
0.338079 |
1
|
1364
|
|
GO:0016020
|
membrane
|
0.342534 |
0.342534 |
1
|
2122
|
|
GO:0046552
|
photoreceptor cell fate commitment
|
0.345238 |
0.345238 |
1
|
58
|
|
GO:0001751
|
compound eye photoreceptor cell differentiation
|
0.354497 |
0.354497 |
1
|
134
|
|
GO:0051321
|
meiotic cell cycle
|
0.357143 |
0.357143 |
1
|
270
|
|
GO:0001754
|
eye photoreceptor cell differentiation
|
0.361386 |
0.361386 |
1
|
146
|
|
GO:0048072
|
compound eye pigmentation
|
0.361702 |
0.361702 |
1
|
17
|
|
GO:0007051
|
spindle organization
|
0.368421 |
0.368421 |
1
|
238
|
|
GO:0005216
|
ion channel activity
|
0.369141 |
0.369141 |
1
|
189
|
|
GO:0042706
|
eye photoreceptor cell fate commitment
|
0.371622 |
0.371622 |
1
|
55
|
|
GO:0009887
|
organ morphogenesis
|
0.374384 |
0.374384 |
1
|
684
|
|
GO:0007444
|
imaginal disc development
|
0.376006 |
0.376006 |
1
|
467
|
|
GO:0051234
|
establishment of localization
|
0.377001 |
0.377001 |
1
|
1549
|
|
GO:0072507
|
divalent inorganic cation homeostasis
|
0.388889 |
0.388889 |
1
|
14
|
|
GO:0007173
|
epidermal growth factor receptor signaling pathway
|
0.407407 |
0.407407 |
1
|
55
|
|
GO:0090066
|
regulation of anatomical structure size
|
0.410151 |
0.410151 |
1
|
169
|
|
GO:0001752
|
compound eye photoreceptor fate commitment
|
0.410448 |
0.410448 |
1
|
55
|
|
GO:0048563
|
post-embryonic organ morphogenesis
|
0.415167 |
0.415167 |
1
|
323
|
|
GO:0023034
|
intracellular signaling pathway
|
0.430962 |
0.430962 |
1
|
412
|
|
GO:0007423
|
sensory organ development
|
0.438003 |
0.438003 |
1
|
544
|
|
GO:0048869
|
cellular developmental process
|
0.442161 |
0.442161 |
1
|
1276
|
|
GO:0030182
|
neuron differentiation
|
0.442292 |
0.442292 |
1
|
548
|
|
GO:0007165
|
signal transduction
|
0.443656 |
0.443656 |
1
|
548
|
|
GO:0003677
|
DNA binding
|
0.444205 |
0.444205 |
1
|
824
|
|
GO:0051179
|
localization
|
0.445834 |
0.445834 |
1
|
1896
|
|
GO:0022008
|
neurogenesis
|
0.452398 |
0.452398 |
1
|
632
|
|
GO:0048468
|
cell development
|
0.457419 |
0.457419 |
1
|
1042
|
|
GO:0048609
|
multicellular organismal reproductive process
|
0.480389 |
0.480389 |
1
|
937
|
|
GO:0032504
|
multicellular organism reproduction
|
0.480389 |
0.480389 |
1
|
937
|
|
GO:0016772
|
transferase activity, transferring phosphorus-containing groups
|
0.480583 |
0.480583 |
1
|
495
|
|
GO:0035556
|
intracellular signal transduction
|
0.499208 |
0.499208 |
1
|
315
|
|
GO:0007399
|
nervous system development
|
0.500000 |
0.500000 |
1
|
848
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.524233 |
0.524233 |
2
|
1517
|
|
GO:0000226
|
microtubule cytoskeleton organization
|
0.529687 |
0.529687 |
1
|
339
|
|
GO:0048592
|
eye morphogenesis
|
0.542069 |
0.542069 |
1
|
393
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0000278
|
mitotic cell cycle
|
0.567460 |
0.567460 |
1
|
429
|
|
GO:0003674
|
molecular_function
|
0.567526 |
0.567526 |
4
|
11064
|
|
GO:0032502
|
developmental process
|
0.575616 |
0.575616 |
1
|
2638
|
|
GO:0007560
|
imaginal disc morphogenesis
|
0.575758 |
0.575758 |
1
|
323
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0006996
|
organelle organization
|
0.606991 |
0.606991 |
2
|
1182
|
|
GO:0042330
|
taxis
|
0.628051 |
0.628051 |
1
|
237
|
|
GO:0055080
|
cation homeostasis
|
0.631579 |
0.631579 |
1
|
36
|
|
GO:0035120
|
post-embryonic appendage morphogenesis
|
0.638095 |
0.638095 |
1
|
268
|
|
GO:0030001
|
metal ion transport
|
0.639676 |
0.639676 |
1
|
158
|
|
GO:0042221
|
response to chemical stimulus
|
0.650010 |
0.650010 |
1
|
632
|
|
GO:0007166
|
cell surface receptor linked signaling pathway
|
0.652720 |
0.652720 |
1
|
624
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0048069
|
eye pigmentation
|
0.661972 |
0.661972 |
1
|
47
|
|
GO:0007472
|
wing disc morphogenesis
|
0.665796 |
0.665796 |
1
|
255
|
|
GO:0055065
|
metal ion homeostasis
|
0.666667 |
0.666667 |
1
|
24
|
|
GO:0035220
|
wing disc development
|
0.680942 |
0.680942 |
1
|
318
|
|
GO:0007169
|
transmembrane receptor protein tyrosine kinase signaling pathway
|
0.685279 |
0.685279 |
1
|
135
|
|
GO:0003676
|
nucleic acid binding
|
0.697754 |
0.697754 |
1
|
1855
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0007010
|
cytoskeleton organization
|
0.701513 |
0.701513 |
1
|
536
|
|
GO:0005623
|
cell
|
0.702620 |
0.702620 |
1
|
6195
|
|
GO:0044464
|
cell part
|
0.702620 |
0.702620 |
1
|
6195
|
|
GO:0007292
|
female gamete generation
|
0.727380 |
0.727380 |
1
|
619
|
|
GO:0004672
|
protein kinase activity
|
0.728643 |
0.728643 |
1
|
290
|
|
GO:0016773
|
phosphotransferase activity, alcohol group as acceptor
|
0.735354 |
0.735354 |
1
|
364
|
|
GO:0006812
|
cation transport
|
0.746224 |
0.746224 |
1
|
247
|
|
GO:0030718
|
germ-line stem cell maintenance
|
0.760870 |
0.760870 |
1
|
35
|
|
GO:0008361
|
regulation of cell size
|
0.769231 |
0.769231 |
1
|
100
|
|
GO:0022891
|
substrate-specific transmembrane transporter activity
|
0.788146 |
0.788146 |
1
|
625
|
|
GO:0016301
|
kinase activity
|
0.795960 |
0.795960 |
1
|
394
|
|
GO:0008324
|
cation transmembrane transporter activity
|
0.808679 |
0.808679 |
1
|
410
|
|
GO:0015075
|
ion transmembrane transporter activity
|
0.811200 |
0.811200 |
1
|
507
|
|
GO:0001745
|
compound eye morphogenesis
|
0.811947 |
0.811947 |
1
|
367
|
|
GO:0022892
|
substrate-specific transporter activity
|
0.815728 |
0.815728 |
1
|
695
|
|
GO:0001654
|
eye development
|
0.841912 |
0.841912 |
1
|
458
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.842767 |
0.842767 |
1
|
402
|
|
GO:0003824
|
catalytic activity
|
0.843631 |
0.843631 |
1
|
4106
|
|
GO:0022857
|
transmembrane transporter activity
|
0.852113 |
0.852113 |
1
|
726
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0019953
|
sexual reproduction
|
0.859100 |
0.859100 |
1
|
878
|
|
GO:0008150
|
biological_process
|
0.867374 |
0.867374 |
3
|
10616
|
|
GO:0007276
|
gamete generation
|
0.883697 |
0.883697 |
1
|
851
|
|
GO:0007275
|
multicellular organismal development
|
0.887792 |
0.887792 |
1
|
2296
|
|
GO:0022403
|
cell cycle phase
|
0.896183 |
0.896183 |
1
|
587
|
|
GO:0007476
|
imaginal disc-derived wing morphogenesis
|
0.903915 |
0.903915 |
1
|
254
|
|
GO:0050801
|
ion homeostasis
|
0.925024 |
0.925024 |
1
|
57
|
|
GO:0048749
|
compound eye development
|
0.932314 |
0.932314 |
1
|
427
|
|
GO:0000279
|
M phase
|
0.945486 |
0.945486 |
1
|
555
|
|
GO:0016043
|
cellular component organization
|
0.947563 |
0.947563 |
2
|
2052
|
|
GO:0002165
|
instar larval or pupal development
|
0.954000 |
0.954000 |
1
|
477
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0035114
|
imaginal disc-derived appendage morphogenesis
|
0.975524 |
0.975524 |
1
|
279
|
|
GO:0048737
|
imaginal disc-derived appendage development
|
0.986014 |
0.986014 |
1
|
282
|
|
GO:0048477
|
oogenesis
|
0.987076 |
0.987076 |
1
|
611
|
|
GO:0005575
|
cellular_component
|
0.990974 |
0.990974 |
1
|
8817
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
4
|
12747
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|
|
GO:0015267
|
channel activity
|
1.00000 |
1.00000 |
1
|
201
|
|
GO:0031667
|
response to nutrient levels
|
1.00000 |
1.00000 |
1
|
36
|