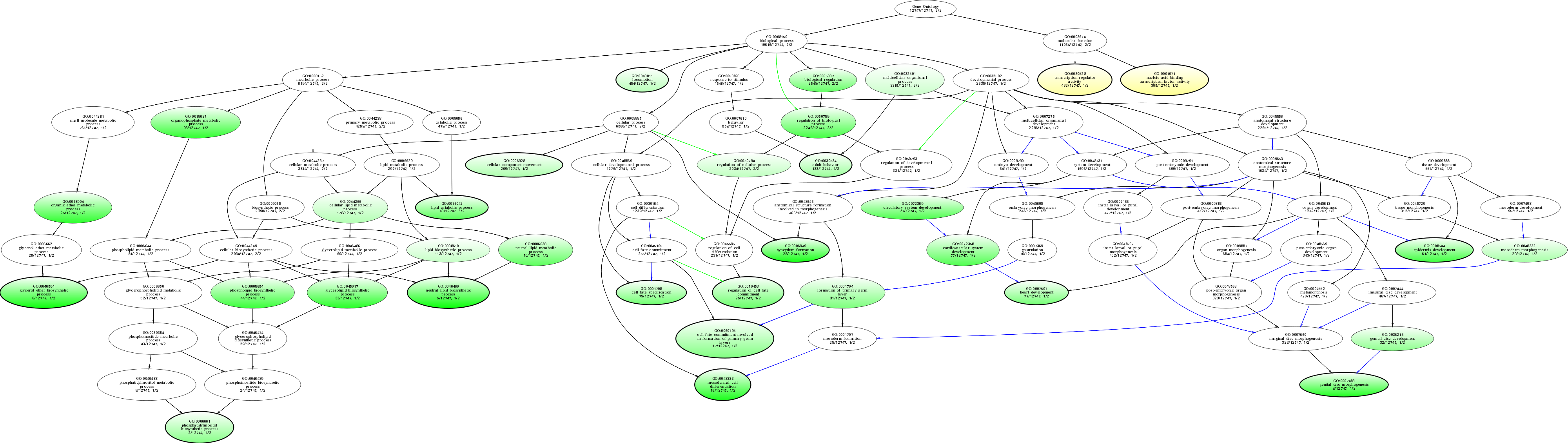
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0046460
|
neutral lipid biosynthetic process
|
0.00489236 |
0.00489236 |
1
|
5
|
|
GO:0046504
|
glycerol ether biosynthetic process
|
0.00584367 |
0.00584367 |
1
|
6
|
|
GO:0006949
|
syncytium formation
|
0.00847903 |
0.00847903 |
1
|
28
|
|
GO:0048333
|
mesodermal cell differentiation
|
0.0128617 |
0.0128617 |
1
|
16
|
|
GO:0007483
|
genital disc morphogenesis
|
0.0269461 |
0.0269461 |
1
|
9
|
|
GO:0045017
|
glycerolipid biosynthetic process
|
0.0317440 |
0.0317440 |
1
|
33
|
|
GO:0018904
|
organic ether metabolic process
|
0.0328515 |
0.0328515 |
1
|
25
|
|
GO:0019637
|
organophosphate metabolic process
|
0.0354933 |
0.0354933 |
1
|
93
|
|
GO:0008654
|
phospholipid biosynthetic process
|
0.0419700 |
0.0419700 |
1
|
44
|
|
GO:0008544
|
epidermis development
|
0.0438533 |
0.0438533 |
1
|
61
|
|
GO:0050789
|
regulation of biological process
|
0.0447451 |
0.0447451 |
2
|
2246
|
|
GO:0072359
|
circulatory system development
|
0.0454009 |
0.0454009 |
1
|
77
|
|
GO:0072358
|
cardiovascular system development
|
0.0454009 |
0.0454009 |
1
|
77
|
|
GO:0016042
|
lipid catabolic process
|
0.0551724 |
0.0551724 |
1
|
40
|
|
GO:0006638
|
neutral lipid metabolic process
|
0.0561798 |
0.0561798 |
1
|
10
|
|
GO:0065007
|
biological regulation
|
0.0575901 |
0.0575901 |
2
|
2548
|
|
GO:0010453
|
regulation of cell fate commitment
|
0.0612745 |
0.0612745 |
1
|
25
|
|
GO:0001708
|
cell fate specification
|
0.0619122 |
0.0619122 |
1
|
79
|
|
GO:0007507
|
heart development
|
0.0619968 |
0.0619968 |
1
|
77
|
|
GO:0001704
|
formation of primary germ layer
|
0.0633947 |
0.0633947 |
1
|
31
|
|
GO:0060795
|
cell fate commitment involved in formation of primary germ layers
|
0.0636704 |
0.0636704 |
1
|
17
|
|
GO:0006661
|
phosphatidylinositol biosynthetic process
|
0.0666667 |
0.0666667 |
1
|
2
|
|
GO:0035215
|
genital disc development
|
0.0685225 |
0.0685225 |
1
|
32
|
|
GO:0001071
|
nucleic acid binding transcription factor activity
|
0.0701313 |
0.0701313 |
1
|
395
|
|
GO:0030534
|
adult behavior
|
0.0762108 |
0.0762108 |
1
|
133
|
|
GO:0030528
|
transcription regulator activity
|
0.0765699 |
0.0765699 |
1
|
432
|
|
GO:0048332
|
mesoderm morphogenesis
|
0.0794521 |
0.0794521 |
1
|
29
|
|
GO:0006928
|
cellular component movement
|
0.0803367 |
0.0803367 |
1
|
269
|
|
GO:0044255
|
cellular lipid metabolic process
|
0.0890763 |
0.0890763 |
1
|
178
|
|
GO:0040011
|
locomotion
|
0.0891086 |
0.0891086 |
1
|
484
|
|
GO:0050794
|
regulation of cellular process
|
0.0938314 |
0.0938314 |
2
|
2034
|
|
GO:0008610
|
lipid biosynthetic process
|
0.0973950 |
0.0973950 |
1
|
113
|
|
GO:0032501
|
multicellular organismal process
|
0.0974889 |
0.0974889 |
2
|
3315
|
|
GO:0042692
|
muscle cell differentiation
|
0.102644 |
0.102644 |
1
|
132
|
|
GO:2000026
|
regulation of multicellular organismal development
|
0.109792 |
0.109792 |
1
|
259
|
|
GO:0061061
|
muscle structure development
|
0.111258 |
0.111258 |
1
|
252
|
|
GO:0007501
|
mesodermal cell fate specification
|
0.130952 |
0.130952 |
1
|
11
|
|
GO:0050767
|
regulation of neurogenesis
|
0.131004 |
0.131004 |
1
|
90
|
|
GO:0046463
|
acylglycerol biosynthetic process
|
0.131579 |
0.131579 |
1
|
5
|
|
GO:0045664
|
regulation of neuron differentiation
|
0.131907 |
0.131907 |
1
|
74
|
|
GO:0051960
|
regulation of nervous system development
|
0.132196 |
0.132196 |
1
|
124
|
|
GO:0008081
|
phosphoric diester hydrolase activity
|
0.132420 |
0.132420 |
1
|
29
|
|
GO:0006629
|
lipid metabolic process
|
0.132436 |
0.132436 |
1
|
292
|
|
GO:0006639
|
acylglycerol metabolic process
|
0.135135 |
0.135135 |
1
|
10
|
|
GO:0007417
|
central nervous system development
|
0.135613 |
0.135613 |
1
|
230
|
|
GO:0004629
|
phospholipase C activity
|
0.136364 |
0.136364 |
1
|
9
|
|
GO:0048598
|
embryonic morphogenesis
|
0.136979 |
0.136979 |
1
|
243
|
|
GO:0023046
|
signaling process
|
0.138061 |
0.138061 |
1
|
760
|
|
GO:0051674
|
localization of cell
|
0.138713 |
0.138713 |
1
|
263
|
|
GO:0005509
|
calcium ion binding
|
0.144237 |
0.144237 |
1
|
209
|
|
GO:0007517
|
muscle organ development
|
0.149102 |
0.149102 |
1
|
191
|
|
GO:0060284
|
regulation of cell development
|
0.159091 |
0.159091 |
1
|
168
|
|
GO:0051239
|
regulation of multicellular organismal process
|
0.161772 |
0.161772 |
1
|
368
|
|
GO:0009058
|
biosynthetic process
|
0.161869 |
0.161869 |
2
|
2090
|
|
GO:0050793
|
regulation of developmental process
|
0.161873 |
0.161873 |
1
|
321
|
|
GO:0060537
|
muscle tissue development
|
0.163375 |
0.163375 |
1
|
91
|
|
GO:0045595
|
regulation of cell differentiation
|
0.167859 |
0.167859 |
1
|
231
|
|
GO:0007498
|
mesoderm development
|
0.170557 |
0.170557 |
1
|
95
|
|
GO:0009056
|
catabolic process
|
0.175955 |
0.175955 |
1
|
479
|
|
GO:0048646
|
anatomical structure formation involved in morphogenesis
|
0.176649 |
0.176649 |
1
|
466
|
|
GO:0046488
|
phosphatidylinositol metabolic process
|
0.186047 |
0.186047 |
1
|
8
|
|
GO:0009791
|
post-embryonic development
|
0.189538 |
0.189538 |
1
|
500
|
|
GO:0048729
|
tissue morphogenesis
|
0.194151 |
0.194151 |
1
|
312
|
|
GO:0016298
|
lipase activity
|
0.195565 |
0.195565 |
1
|
97
|
|
GO:0045165
|
cell fate commitment
|
0.199843 |
0.199843 |
1
|
255
|
|
GO:0014902
|
myotube differentiation
|
0.225806 |
0.225806 |
1
|
28
|
|
GO:0008152
|
metabolic process
|
0.239353 |
0.239353 |
2
|
5194
|
|
GO:0023052
|
signaling
|
0.240473 |
0.240473 |
1
|
1364
|
|
GO:0044237
|
cellular metabolic process
|
0.241038 |
0.241038 |
2
|
3814
|
|
GO:0009790
|
embryo development
|
0.242987 |
0.242987 |
1
|
641
|
|
GO:0009888
|
tissue development
|
0.245916 |
0.245916 |
1
|
557
|
|
GO:0016788
|
hydrolase activity, acting on ester bonds
|
0.251521 |
0.251521 |
1
|
496
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.259446 |
0.259446 |
1
|
412
|
|
GO:0005488
|
binding
|
0.259926 |
0.259926 |
2
|
5641
|
|
GO:0050896
|
response to stimulus
|
0.270384 |
0.270384 |
1
|
1548
|
|
GO:0044281
|
small molecule metabolic process
|
0.271588 |
0.271588 |
1
|
761
|
|
GO:0007552
|
metamorphosis
|
0.273794 |
0.273794 |
1
|
420
|
|
GO:0048569
|
post-embryonic organ development
|
0.276167 |
0.276167 |
1
|
343
|
|
GO:0044249
|
cellular biosynthetic process
|
0.277461 |
0.277461 |
2
|
2034
|
|
GO:0007626
|
locomotory behavior
|
0.280859 |
0.280859 |
1
|
157
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.283034 |
0.283034 |
1
|
1056
|
|
GO:0003704
|
specific RNA polymerase II transcription factor activity
|
0.292135 |
0.292135 |
1
|
78
|
|
GO:0046339
|
diacylglycerol metabolic process
|
0.300000 |
0.300000 |
1
|
3
|
|
GO:0042659
|
regulation of cell fate specification
|
0.300000 |
0.300000 |
1
|
24
|
|
GO:0043565
|
sequence-specific DNA binding
|
0.300971 |
0.300971 |
1
|
248
|
|
GO:0007369
|
gastrulation
|
0.312757 |
0.312757 |
1
|
76
|
|
GO:0048869
|
cellular developmental process
|
0.322329 |
0.322329 |
1
|
1276
|
|
GO:0051179
|
localization
|
0.325313 |
0.325313 |
1
|
1896
|
|
GO:0004435
|
phosphoinositide phospholipase C activity
|
0.333333 |
0.333333 |
1
|
3
|
|
GO:0046486
|
glycerolipid metabolic process
|
0.337079 |
0.337079 |
1
|
60
|
|
GO:0007610
|
behavior
|
0.361111 |
0.361111 |
1
|
559
|
|
GO:0009887
|
organ morphogenesis
|
0.374384 |
0.374384 |
1
|
684
|
|
GO:0007444
|
imaginal disc development
|
0.376006 |
0.376006 |
1
|
467
|
|
GO:0019222
|
regulation of metabolic process
|
0.376562 |
0.376562 |
1
|
1242
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.381695 |
0.381695 |
1
|
905
|
|
GO:0009987
|
cellular process
|
0.381822 |
0.381822 |
2
|
6560
|
|
GO:0006350
|
transcription
|
0.387810 |
0.387810 |
1
|
859
|
|
GO:0043226
|
organelle
|
0.401157 |
0.401157 |
1
|
3537
|
|
GO:0016070
|
RNA metabolic process
|
0.413829 |
0.413829 |
1
|
1191
|
|
GO:0048563
|
post-embryonic organ morphogenesis
|
0.415167 |
0.415167 |
1
|
323
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.418200 |
0.418200 |
1
|
1039
|
|
GO:0046474
|
glycerophospholipid biosynthetic process
|
0.420290 |
0.420290 |
1
|
29
|
|
GO:0006644
|
phospholipid metabolic process
|
0.428571 |
0.428571 |
1
|
81
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.429576 |
0.429576 |
1
|
1110
|
|
GO:0023034
|
intracellular signaling pathway
|
0.430962 |
0.430962 |
1
|
412
|
|
GO:0032502
|
developmental process
|
0.435255 |
0.435255 |
1
|
2638
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.440308 |
0.440308 |
1
|
686
|
|
GO:0042578
|
phosphoric ester hydrolase activity
|
0.441532 |
0.441532 |
1
|
219
|
|
GO:0030182
|
neuron differentiation
|
0.442292 |
0.442292 |
1
|
548
|
|
GO:0007165
|
signal transduction
|
0.443656 |
0.443656 |
1
|
548
|
|
GO:0003677
|
DNA binding
|
0.444205 |
0.444205 |
1
|
824
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.451726 |
0.451726 |
1
|
903
|
|
GO:0022008
|
neurogenesis
|
0.452398 |
0.452398 |
1
|
632
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.456989 |
0.456989 |
1
|
850
|
|
GO:0048468
|
cell development
|
0.457419 |
0.457419 |
1
|
1042
|
|
GO:0043167
|
ion binding
|
0.460626 |
0.460626 |
1
|
1498
|
|
GO:0016787
|
hydrolase activity
|
0.480273 |
0.480273 |
1
|
1972
|
|
GO:0010468
|
regulation of gene expression
|
0.489930 |
0.489930 |
1
|
973
|
|
GO:0032774
|
RNA biosynthetic process
|
0.496008 |
0.496008 |
1
|
703
|
|
GO:0046489
|
phosphoinositide biosynthetic process
|
0.500000 |
0.500000 |
1
|
24
|
|
GO:0007629
|
flight behavior
|
0.500000 |
0.500000 |
1
|
47
|
|
GO:0007399
|
nervous system development
|
0.500000 |
0.500000 |
1
|
848
|
|
GO:0008344
|
adult locomotory behavior
|
0.502674 |
0.502674 |
1
|
94
|
|
GO:0048870
|
cell motility
|
0.512097 |
0.512097 |
1
|
254
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.513595 |
0.513595 |
1
|
850
|
|
GO:0010467
|
gene expression
|
0.531494 |
0.531494 |
1
|
1907
|
|
GO:0060538
|
skeletal muscle organ development
|
0.539267 |
0.539267 |
1
|
103
|
|
GO:0004620
|
phospholipase activity
|
0.546392 |
0.546392 |
1
|
53
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0003676
|
nucleic acid binding
|
0.549587 |
0.549587 |
1
|
1855
|
|
GO:0007560
|
imaginal disc morphogenesis
|
0.575758 |
0.575758 |
1
|
323
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0006650
|
glycerophospholipid metabolic process
|
0.584270 |
0.584270 |
1
|
52
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.599142 |
0.599142 |
1
|
888
|
|
GO:0006651
|
diacylglycerol biosynthetic process
|
0.600000 |
0.600000 |
1
|
3
|
|
GO:0003824
|
catalytic activity
|
0.604523 |
0.604523 |
1
|
4106
|
|
GO:0003702
|
RNA polymerase II transcription factor activity
|
0.618056 |
0.618056 |
1
|
267
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.635595 |
0.635595 |
1
|
888
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.643595 |
0.643595 |
1
|
2093
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.646807 |
0.646807 |
1
|
1623
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.658716 |
0.658716 |
1
|
1789
|
|
GO:0007519
|
skeletal muscle tissue development
|
0.669565 |
0.669565 |
1
|
77
|
|
GO:0044238
|
primary metabolic process
|
0.672346 |
0.672346 |
2
|
4259
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.684327 |
0.684327 |
1
|
620
|
|
GO:0045449
|
regulation of transcription
|
0.687166 |
0.687166 |
1
|
771
|
|
GO:0008150
|
biological_process
|
0.693584 |
0.693584 |
2
|
10616
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0005623
|
cell
|
0.702620 |
0.702620 |
1
|
6195
|
|
GO:0044464
|
cell part
|
0.702620 |
0.702620 |
1
|
6195
|
|
GO:0044424
|
intracellular part
|
0.707667 |
0.707667 |
1
|
4384
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.746994 |
0.746994 |
1
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.748407 |
0.748407 |
1
|
1959
|
|
GO:0003674
|
molecular_function
|
0.753361 |
0.753361 |
2
|
11064
|
|
GO:0005622
|
intracellular
|
0.764165 |
0.764165 |
1
|
4734
|
|
GO:0043229
|
intracellular organelle
|
0.804100 |
0.804100 |
1
|
3530
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.814170 |
0.814170 |
1
|
701
|
|
GO:0030384
|
phosphoinositide metabolic process
|
0.826923 |
0.826923 |
1
|
43
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.842767 |
0.842767 |
1
|
402
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.869824 |
0.869824 |
1
|
2878
|
|
GO:0001707
|
mesoderm formation
|
0.875000 |
0.875000 |
1
|
28
|
|
GO:0007275
|
multicellular organismal development
|
0.887792 |
0.887792 |
1
|
2296
|
|
GO:0043170
|
macromolecule metabolic process
|
0.904435 |
0.904435 |
1
|
3588
|
|
GO:0005575
|
cellular_component
|
0.904963 |
0.904963 |
1
|
8817
|
|
GO:0051146
|
striated muscle cell differentiation
|
0.939394 |
0.939394 |
1
|
124
|
|
GO:0001710
|
mesodermal cell fate commitment
|
0.941176 |
0.941176 |
1
|
16
|
|
GO:0002165
|
instar larval or pupal development
|
0.954000 |
0.954000 |
1
|
477
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0016477
|
cell migration
|
0.964567 |
0.964567 |
1
|
245
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0014706
|
striated muscle tissue development
|
0.989011 |
0.989011 |
1
|
90
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
2
|
12747
|
|
GO:0000768
|
syncytium formation by plasma membrane fusion
|
1.00000 |
1.00000 |
1
|
28
|
|
GO:0003700
|
sequence-specific DNA binding transcription factor activity
|
1.00000 |
1.00000 |
1
|
395
|
|
GO:0006662
|
glycerol ether metabolic process
|
1.00000 |
1.00000 |
1
|
25
|
|
GO:0007520
|
myoblast fusion
|
1.00000 |
1.00000 |
1
|
28
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|