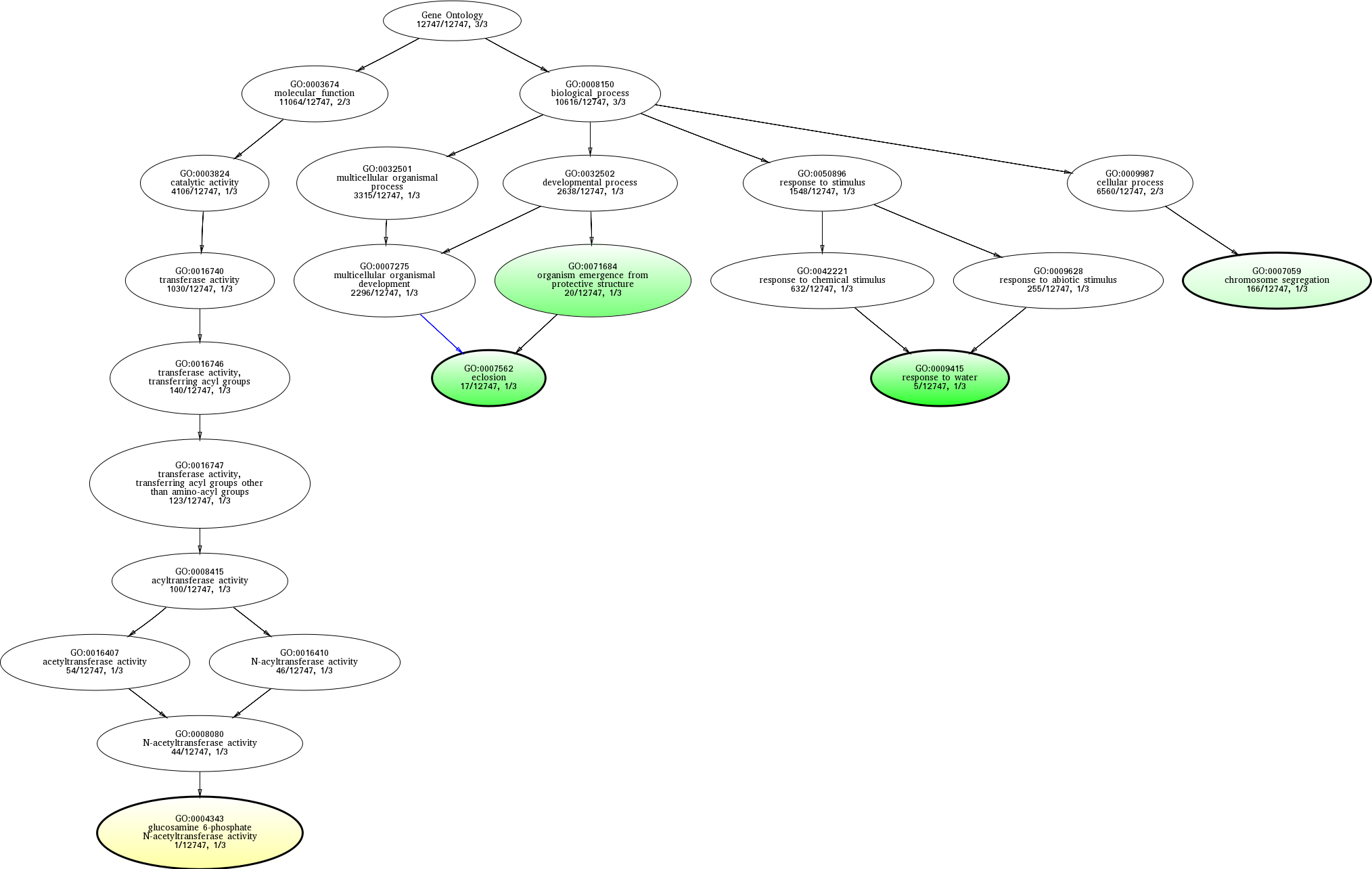
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0009415
|
response to water
|
0.00595238 |
0.00595238 |
1
|
5
|
|
GO:0007562
|
eclosion
|
0.00740418 |
0.00740418 |
1
|
17
|
|
GO:0071684
|
organism emergence from protective structure
|
0.00758150 |
0.00758150 |
1
|
20
|
|
GO:0004343
|
glucosamine 6-phosphate N-acetyltransferase activity
|
0.0227273 |
0.0227273 |
1
|
1
|
|
GO:0007059
|
chromosome segregation
|
0.0499732 |
0.0499732 |
1
|
166
|
|
GO:0016321
|
female meiosis chromosome segregation
|
0.103817 |
0.103817 |
1
|
68
|
|
GO:0055085
|
transmembrane transport
|
0.132219 |
0.132219 |
1
|
469
|
|
GO:0016746
|
transferase activity, transferring acyl groups
|
0.135922 |
0.135922 |
1
|
140
|
|
GO:0005215
|
transporter activity
|
0.148089 |
0.148089 |
1
|
852
|
|
GO:0072511
|
divalent inorganic cation transport
|
0.149798 |
0.149798 |
1
|
37
|
|
GO:0009628
|
response to abiotic stimulus
|
0.164729 |
0.164729 |
1
|
255
|
|
GO:0022402
|
cell cycle process
|
0.189739 |
0.189739 |
1
|
655
|
|
GO:0007049
|
cell cycle
|
0.217222 |
0.217222 |
1
|
756
|
|
GO:0005262
|
calcium channel activity
|
0.218750 |
0.218750 |
1
|
28
|
|
GO:0006811
|
ion transport
|
0.221109 |
0.221109 |
1
|
331
|
|
GO:0070838
|
divalent metal ion transport
|
0.227848 |
0.227848 |
1
|
36
|
|
GO:0016740
|
transferase activity
|
0.250852 |
0.250852 |
1
|
1030
|
|
GO:0005261
|
cation channel activity
|
0.272340 |
0.272340 |
1
|
128
|
|
GO:0022803
|
passive transmembrane transporter activity
|
0.276860 |
0.276860 |
1
|
201
|
|
GO:0022838
|
substrate-specific channel activity
|
0.307448 |
0.307448 |
1
|
194
|
|
GO:0045132
|
meiotic chromosome segregation
|
0.315287 |
0.315287 |
1
|
99
|
|
GO:0016020
|
membrane
|
0.342534 |
0.342534 |
1
|
2122
|
|
GO:0051321
|
meiotic cell cycle
|
0.357143 |
0.357143 |
1
|
270
|
|
GO:0005216
|
ion channel activity
|
0.369141 |
0.369141 |
1
|
189
|
|
GO:0050896
|
response to stimulus
|
0.376795 |
0.376795 |
1
|
1548
|
|
GO:0051234
|
establishment of localization
|
0.377001 |
0.377001 |
1
|
1549
|
|
GO:0009270
|
response to humidity
|
0.400000 |
0.400000 |
1
|
2
|
|
GO:0007143
|
female meiosis
|
0.401575 |
0.401575 |
1
|
102
|
|
GO:0042221
|
response to chemical stimulus
|
0.408269 |
0.408269 |
1
|
632
|
|
GO:0007126
|
meiosis
|
0.432709 |
0.432709 |
1
|
254
|
|
GO:0051179
|
localization
|
0.445834 |
0.445834 |
1
|
1896
|
|
GO:0051327
|
M phase of meiotic cell cycle
|
0.456014 |
0.456014 |
1
|
254
|
|
GO:0016410
|
N-acyltransferase activity
|
0.460000 |
0.460000 |
1
|
46
|
|
GO:0007399
|
nervous system development
|
0.500000 |
0.500000 |
1
|
848
|
|
GO:0016407
|
acetyltransferase activity
|
0.540000 |
0.540000 |
1
|
54
|
|
GO:0032502
|
developmental process
|
0.575616 |
0.575616 |
1
|
2638
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0003824
|
catalytic activity
|
0.604523 |
0.604523 |
1
|
4106
|
|
GO:0030001
|
metal ion transport
|
0.639676 |
0.639676 |
1
|
158
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0009987
|
cellular process
|
0.673638 |
0.673638 |
2
|
6560
|
|
GO:0032501
|
multicellular organismal process
|
0.674757 |
0.674757 |
1
|
3315
|
|
GO:0005623
|
cell
|
0.702620 |
0.702620 |
1
|
6195
|
|
GO:0044464
|
cell part
|
0.702620 |
0.702620 |
1
|
6195
|
|
GO:0006812
|
cation transport
|
0.746224 |
0.746224 |
1
|
247
|
|
GO:0008080
|
N-acetyltransferase activity
|
0.785714 |
0.785714 |
1
|
44
|
|
GO:0022891
|
substrate-specific transmembrane transporter activity
|
0.788146 |
0.788146 |
1
|
625
|
|
GO:0008324
|
cation transmembrane transporter activity
|
0.808679 |
0.808679 |
1
|
410
|
|
GO:0015075
|
ion transmembrane transporter activity
|
0.811200 |
0.811200 |
1
|
507
|
|
GO:0008415
|
acyltransferase activity
|
0.813008 |
0.813008 |
1
|
100
|
|
GO:0022892
|
substrate-specific transporter activity
|
0.815728 |
0.815728 |
1
|
695
|
|
GO:0022857
|
transmembrane transporter activity
|
0.852113 |
0.852113 |
1
|
726
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0008152
|
metabolic process
|
0.866808 |
0.866808 |
1
|
5194
|
|
GO:0016747
|
transferase activity, transferring acyl groups other than amino-acyl groups
|
0.878571 |
0.878571 |
1
|
123
|
|
GO:0006816
|
calcium ion transport
|
0.888889 |
0.888889 |
1
|
32
|
|
GO:0022403
|
cell cycle phase
|
0.896183 |
0.896183 |
1
|
587
|
|
GO:0000279
|
M phase
|
0.945486 |
0.945486 |
1
|
555
|
|
GO:0003674
|
molecular_function
|
0.952326 |
0.952326 |
2
|
11064
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0005575
|
cellular_component
|
0.970710 |
0.970710 |
1
|
8817
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0015267
|
channel activity
|
1.00000 |
1.00000 |
1
|
201
|