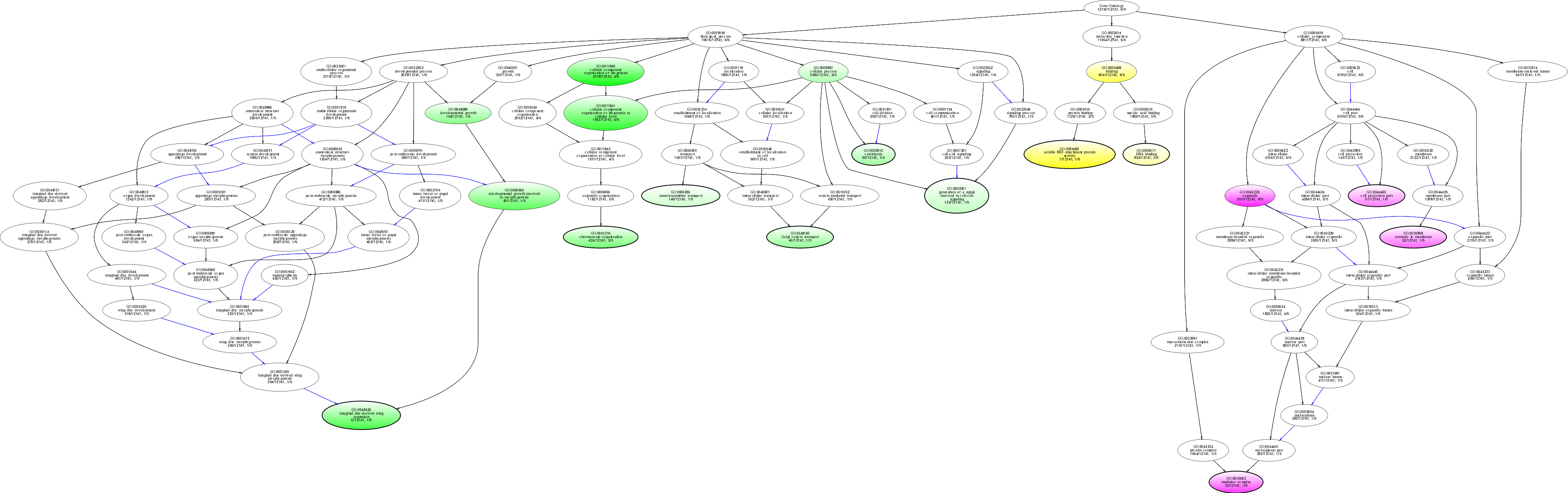
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.00652549 |
0.00652549 |
4
|
2108
|
|
GO:0005483
|
soluble NSF attachment protein activity
|
0.00809713 |
0.00809713 |
1
|
7
|
|
GO:0043226
|
organelle
|
0.0103714 |
0.0103714 |
5
|
3537
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.0128307 |
0.0128307 |
4
|
1562
|
|
GO:0016592
|
mediator complex
|
0.0204588 |
0.0204588 |
1
|
33
|
|
GO:0048526
|
imaginal disc-derived wing expansion
|
0.0206186 |
0.0206186 |
1
|
6
|
|
GO:0005488
|
binding
|
0.0344225 |
0.0344225 |
5
|
5641
|
|
GO:0060560
|
developmental growth involved in morphogenesis
|
0.0355105 |
0.0355105 |
1
|
56
|
|
GO:0019898
|
extrinsic to membrane
|
0.0371960 |
0.0371960 |
1
|
52
|
|
GO:0051276
|
chromosome organization
|
0.0459483 |
0.0459483 |
3
|
424
|
|
GO:0044463
|
cell projection part
|
0.0482885 |
0.0482885 |
1
|
61
|
|
GO:0048589
|
developmental growth
|
0.0576779 |
0.0576779 |
1
|
154
|
|
GO:0048193
|
Golgi vesicle transport
|
0.0693816 |
0.0693816 |
1
|
46
|
|
GO:0000910
|
cytokinesis
|
0.0710813 |
0.0710813 |
1
|
96
|
|
GO:0003677
|
DNA binding
|
0.0874722 |
0.0874722 |
3
|
824
|
|
GO:0009987
|
cellular process
|
0.0900454 |
0.0900454 |
5
|
6560
|
|
GO:0003001
|
generation of a signal involved in cell-cell signaling
|
0.0917389 |
0.0917389 |
1
|
125
|
|
GO:0006836
|
neurotransmitter transport
|
0.0975284 |
0.0975284 |
1
|
146
|
|
GO:0046903
|
secretion
|
0.101536 |
0.101536 |
1
|
152
|
|
GO:0032940
|
secretion by cell
|
0.104981 |
0.104981 |
1
|
144
|
|
GO:0006352
|
transcription initiation, DNA-dependent
|
0.108108 |
0.108108 |
1
|
76
|
|
GO:0042995
|
cell projection
|
0.110243 |
0.110243 |
1
|
143
|
|
GO:0048736
|
appendage development
|
0.110896 |
0.110896 |
1
|
286
|
|
GO:0040007
|
growth
|
0.111065 |
0.111065 |
1
|
247
|
|
GO:0016082
|
synaptic vesicle priming
|
0.121429 |
0.121429 |
1
|
17
|
|
GO:0051301
|
cell division
|
0.147498 |
0.147498 |
1
|
206
|
|
GO:0005575
|
cellular_component
|
0.158275 |
0.158275 |
5
|
8817
|
|
GO:0023061
|
signal release
|
0.164474 |
0.164474 |
1
|
125
|
|
GO:0001505
|
regulation of neurotransmitter levels
|
0.165810 |
0.165810 |
1
|
129
|
|
GO:0070271
|
protein complex biogenesis
|
0.166139 |
0.166139 |
1
|
105
|
|
GO:0006887
|
exocytosis
|
0.167038 |
0.167038 |
1
|
75
|
|
GO:0045184
|
establishment of protein localization
|
0.168623 |
0.168623 |
1
|
273
|
|
GO:0005623
|
cell
|
0.171157 |
0.171157 |
5
|
6195
|
|
GO:0044464
|
cell part
|
0.171157 |
0.171157 |
5
|
6195
|
|
GO:0015031
|
protein transport
|
0.177333 |
0.177333 |
1
|
266
|
|
GO:0044424
|
intracellular part
|
0.177360 |
0.177360 |
5
|
4384
|
|
GO:0005794
|
Golgi apparatus
|
0.178532 |
0.178532 |
1
|
138
|
|
GO:0030528
|
transcription regulator activity
|
0.180596 |
0.180596 |
1
|
432
|
|
GO:0035107
|
appendage morphogenesis
|
0.184365 |
0.184365 |
1
|
283
|
|
GO:0007267
|
cell-cell signaling
|
0.188801 |
0.188801 |
1
|
263
|
|
GO:0009791
|
post-embryonic development
|
0.189538 |
0.189538 |
1
|
500
|
|
GO:0019226
|
transmission of nerve impulse
|
0.194826 |
0.194826 |
1
|
241
|
|
GO:0003008
|
system process
|
0.200302 |
0.200302 |
1
|
664
|
|
GO:0003676
|
nucleic acid binding
|
0.203180 |
0.203180 |
3
|
1855
|
|
GO:0019222
|
regulation of metabolic process
|
0.210401 |
0.210401 |
1
|
1242
|
|
GO:0044306
|
neuron projection terminus
|
0.223301 |
0.223301 |
1
|
23
|
|
GO:0006333
|
chromatin assembly or disassembly
|
0.227304 |
0.227304 |
3
|
167
|
|
GO:0046907
|
intracellular transport
|
0.227832 |
0.227832 |
1
|
352
|
|
GO:0016044
|
cellular membrane organization
|
0.236166 |
0.236166 |
1
|
344
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.237215 |
0.237215 |
1
|
1039
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.244709 |
0.244709 |
1
|
1110
|
|
GO:0048489
|
synaptic vesicle transport
|
0.246512 |
0.246512 |
1
|
106
|
|
GO:0005783
|
endoplasmic reticulum
|
0.248828 |
0.248828 |
1
|
199
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.259446 |
0.259446 |
1
|
412
|
|
GO:0005622
|
intracellular
|
0.260446 |
0.260446 |
5
|
4734
|
|
GO:0006367
|
transcription initiation from RNA polymerase II promoter
|
0.261538 |
0.261538 |
1
|
68
|
|
GO:0006325
|
chromatin organization
|
0.265878 |
0.265878 |
3
|
273
|
|
GO:0007552
|
metamorphosis
|
0.273794 |
0.273794 |
1
|
420
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.274620 |
0.274620 |
4
|
1517
|
|
GO:0048569
|
post-embryonic organ development
|
0.276167 |
0.276167 |
1
|
343
|
|
GO:0033036
|
macromolecule localization
|
0.276371 |
0.276371 |
1
|
524
|
|
GO:0016192
|
vesicle-mediated transport
|
0.276860 |
0.276860 |
1
|
430
|
|
GO:0006357
|
regulation of transcription from RNA polymerase II promoter
|
0.282934 |
0.282934 |
1
|
189
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.283034 |
0.283034 |
1
|
1056
|
|
GO:0034622
|
cellular macromolecular complex assembly
|
0.286645 |
0.286645 |
1
|
176
|
|
GO:0032774
|
RNA biosynthetic process
|
0.290017 |
0.290017 |
1
|
703
|
|
GO:0043233
|
organelle lumen
|
0.291360 |
0.291360 |
1
|
634
|
|
GO:0070013
|
intracellular organelle lumen
|
0.293247 |
0.293247 |
1
|
634
|
|
GO:0033267
|
axon part
|
0.301205 |
0.301205 |
1
|
25
|
|
GO:0065003
|
macromolecular complex assembly
|
0.301303 |
0.301303 |
1
|
185
|
|
GO:0051649
|
establishment of localization in cell
|
0.303951 |
0.303951 |
1
|
500
|
|
GO:0007154
|
cell communication
|
0.305417 |
0.305417 |
1
|
461
|
|
GO:0023046
|
signaling process
|
0.310289 |
0.310289 |
1
|
760
|
|
GO:0044451
|
nucleoplasm part
|
0.313253 |
0.313253 |
1
|
260
|
|
GO:0031974
|
membrane-enclosed lumen
|
0.316927 |
0.316927 |
1
|
647
|
|
GO:0016455
|
RNA polymerase II transcription mediator activity
|
0.333333 |
0.333333 |
1
|
32
|
|
GO:0043229
|
intracellular organelle
|
0.335977 |
0.335977 |
5
|
3530
|
|
GO:0005654
|
nucleoplasm
|
0.340964 |
0.340964 |
1
|
283
|
|
GO:0043227
|
membrane-bounded organelle
|
0.344829 |
0.344829 |
5
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.345459 |
0.345459 |
5
|
2856
|
|
GO:0065007
|
biological regulation
|
0.346129 |
0.346129 |
2
|
2548
|
|
GO:0016251
|
general RNA polymerase II transcription factor activity
|
0.359551 |
0.359551 |
1
|
96
|
|
GO:0006366
|
transcription from RNA polymerase II promoter
|
0.360913 |
0.360913 |
1
|
253
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.366791 |
0.366791 |
1
|
888
|
|
GO:0051641
|
cellular localization
|
0.370663 |
0.370663 |
1
|
607
|
|
GO:0009887
|
organ morphogenesis
|
0.374384 |
0.374384 |
1
|
684
|
|
GO:0007444
|
imaginal disc development
|
0.376006 |
0.376006 |
1
|
467
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.381695 |
0.381695 |
1
|
905
|
|
GO:0006350
|
transcription
|
0.387810 |
0.387810 |
1
|
859
|
|
GO:0043933
|
macromolecular complex subunit organization
|
0.392208 |
0.392208 |
1
|
240
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.396252 |
0.396252 |
1
|
888
|
|
GO:0008150
|
biological_process
|
0.400586 |
0.400586 |
5
|
10616
|
|
GO:0009058
|
biosynthetic process
|
0.402387 |
0.402387 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.402965 |
0.402965 |
1
|
2093
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.405649 |
0.405649 |
1
|
1623
|
|
GO:0005634
|
nucleus
|
0.408003 |
0.408003 |
4
|
1826
|
|
GO:0070727
|
cellular macromolecule localization
|
0.408300 |
0.408300 |
1
|
305
|
|
GO:0006888
|
ER to Golgi vesicle-mediated transport
|
0.413043 |
0.413043 |
1
|
19
|
|
GO:0016070
|
RNA metabolic process
|
0.413829 |
0.413829 |
1
|
1191
|
|
GO:0048563
|
post-embryonic organ morphogenesis
|
0.415167 |
0.415167 |
1
|
323
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.415756 |
0.415756 |
1
|
1789
|
|
GO:0016079
|
synaptic vesicle exocytosis
|
0.431507 |
0.431507 |
1
|
63
|
|
GO:0007423
|
sensory organ development
|
0.438003 |
0.438003 |
1
|
544
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.440308 |
0.440308 |
1
|
686
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.451726 |
0.451726 |
1
|
903
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.456989 |
0.456989 |
1
|
850
|
|
GO:0031981
|
nuclear lumen
|
0.475277 |
0.475277 |
1
|
471
|
|
GO:0034613
|
cellular protein localization
|
0.478936 |
0.478936 |
1
|
216
|
|
GO:0006461
|
protein complex assembly
|
0.483871 |
0.483871 |
1
|
105
|
|
GO:0005515
|
protein binding
|
0.484079 |
0.484079 |
2
|
1726
|
|
GO:0034621
|
cellular macromolecular complex subunit organization
|
0.484306 |
0.484306 |
1
|
232
|
|
GO:0010468
|
regulation of gene expression
|
0.489930 |
0.489930 |
1
|
973
|
|
GO:0065008
|
regulation of biological quality
|
0.490437 |
0.490437 |
1
|
729
|
|
GO:0003674
|
molecular_function
|
0.492571 |
0.492571 |
5
|
11064
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.496927 |
0.496927 |
1
|
1617
|
|
GO:0023052
|
signaling
|
0.497295 |
0.497295 |
1
|
1364
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.498346 |
0.498346 |
1
|
1959
|
|
GO:0043005
|
neuron projection
|
0.503497 |
0.503497 |
1
|
72
|
|
GO:0006886
|
intracellular protein transport
|
0.504878 |
0.504878 |
1
|
207
|
|
GO:0043623
|
cellular protein complex assembly
|
0.508108 |
0.508108 |
1
|
94
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.513595 |
0.513595 |
1
|
850
|
|
GO:0061024
|
membrane organization
|
0.521406 |
0.521406 |
1
|
345
|
|
GO:0044249
|
cellular biosynthetic process
|
0.526807 |
0.526807 |
1
|
2034
|
|
GO:0010467
|
gene expression
|
0.531494 |
0.531494 |
1
|
1907
|
|
GO:0048592
|
eye morphogenesis
|
0.542069 |
0.542069 |
1
|
393
|
|
GO:0051234
|
establishment of localization
|
0.545594 |
0.545594 |
1
|
1549
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0071822
|
protein complex subunit organization
|
0.570833 |
0.570833 |
1
|
137
|
|
GO:0007560
|
imaginal disc morphogenesis
|
0.575758 |
0.575758 |
1
|
323
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0003702
|
RNA polymerase II transcription factor activity
|
0.618056 |
0.618056 |
1
|
267
|
|
GO:0051179
|
localization
|
0.626157 |
0.626157 |
1
|
1896
|
|
GO:0035120
|
post-embryonic appendage morphogenesis
|
0.638095 |
0.638095 |
1
|
268
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.639129 |
0.639129 |
1
|
2878
|
|
GO:0006897
|
endocytosis
|
0.655814 |
0.655814 |
1
|
282
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0007472
|
wing disc morphogenesis
|
0.665796 |
0.665796 |
1
|
255
|
|
GO:0006911
|
phagocytosis, engulfment
|
0.680851 |
0.680851 |
1
|
192
|
|
GO:0035220
|
wing disc development
|
0.680942 |
0.680942 |
1
|
318
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.684327 |
0.684327 |
1
|
620
|
|
GO:0045449
|
regulation of transcription
|
0.687166 |
0.687166 |
1
|
771
|
|
GO:0043170
|
macromolecule metabolic process
|
0.690797 |
0.690797 |
1
|
3588
|
|
GO:0030424
|
axon
|
0.694444 |
0.694444 |
1
|
50
|
|
GO:0050789
|
regulation of biological process
|
0.695412 |
0.695412 |
1
|
2246
|
|
GO:0007269
|
neurotransmitter secretion
|
0.701149 |
0.701149 |
1
|
122
|
|
GO:0044428
|
nuclear part
|
0.718426 |
0.718426 |
1
|
830
|
|
GO:0044425
|
membrane part
|
0.721748 |
0.721748 |
1
|
1398
|
|
GO:0006909
|
phagocytosis
|
0.723404 |
0.723404 |
1
|
204
|
|
GO:0022607
|
cellular component assembly
|
0.724655 |
0.724655 |
1
|
577
|
|
GO:0008104
|
protein localization
|
0.727099 |
0.727099 |
1
|
381
|
|
GO:0043234
|
protein complex
|
0.727104 |
0.727104 |
1
|
1564
|
|
GO:0032991
|
macromolecular complex
|
0.753076 |
0.753076 |
1
|
2151
|
|
GO:0044422
|
organelle part
|
0.757673 |
0.757673 |
1
|
2176
|
|
GO:0044085
|
cellular component biogenesis
|
0.759933 |
0.759933 |
1
|
632
|
|
GO:0032502
|
developmental process
|
0.760376 |
0.760376 |
1
|
2638
|
|
GO:0006996
|
organelle organization
|
0.786601 |
0.786601 |
3
|
1182
|
|
GO:0001745
|
compound eye morphogenesis
|
0.811947 |
0.811947 |
1
|
367
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.814170 |
0.814170 |
1
|
701
|
|
GO:0010324
|
membrane invagination
|
0.819767 |
0.819767 |
1
|
282
|
|
GO:0044238
|
primary metabolic process
|
0.819985 |
0.819985 |
1
|
4259
|
|
GO:0050794
|
regulation of cellular process
|
0.839548 |
0.839548 |
1
|
2034
|
|
GO:0001654
|
eye development
|
0.841912 |
0.841912 |
1
|
458
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.842767 |
0.842767 |
1
|
402
|
|
GO:0071844
|
cellular component assembly at cellular level
|
0.846003 |
0.846003 |
1
|
568
|
|
GO:0032501
|
multicellular organismal process
|
0.846212 |
0.846212 |
1
|
3315
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0007268
|
synaptic transmission
|
0.866667 |
0.866667 |
1
|
234
|
|
GO:0016020
|
membrane
|
0.877256 |
0.877256 |
1
|
2122
|
|
GO:0016043
|
cellular component organization
|
0.897828 |
0.897828 |
4
|
2052
|
|
GO:0007476
|
imaginal disc-derived wing morphogenesis
|
0.903915 |
0.903915 |
1
|
254
|
|
GO:0043679
|
axon terminus
|
0.920000 |
0.920000 |
1
|
23
|
|
GO:0048749
|
compound eye development
|
0.932314 |
0.932314 |
1
|
427
|
|
GO:0044444
|
cytoplasmic part
|
0.937226 |
0.937226 |
1
|
1863
|
|
GO:0050877
|
neurological system process
|
0.953313 |
0.953313 |
1
|
633
|
|
GO:0002165
|
instar larval or pupal development
|
0.954000 |
0.954000 |
1
|
477
|
|
GO:0008152
|
metabolic process
|
0.965278 |
0.965278 |
1
|
5194
|
|
GO:0044237
|
cellular metabolic process
|
0.965873 |
0.965873 |
1
|
3814
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0044446
|
intracellular organelle part
|
0.966589 |
0.966589 |
1
|
2162
|
|
GO:0035114
|
imaginal disc-derived appendage morphogenesis
|
0.975524 |
0.975524 |
1
|
279
|
|
GO:0005737
|
cytoplasm
|
0.979996 |
0.979996 |
1
|
2378
|
|
GO:0048737
|
imaginal disc-derived appendage development
|
0.986014 |
0.986014 |
1
|
282
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
5
|
12747
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|