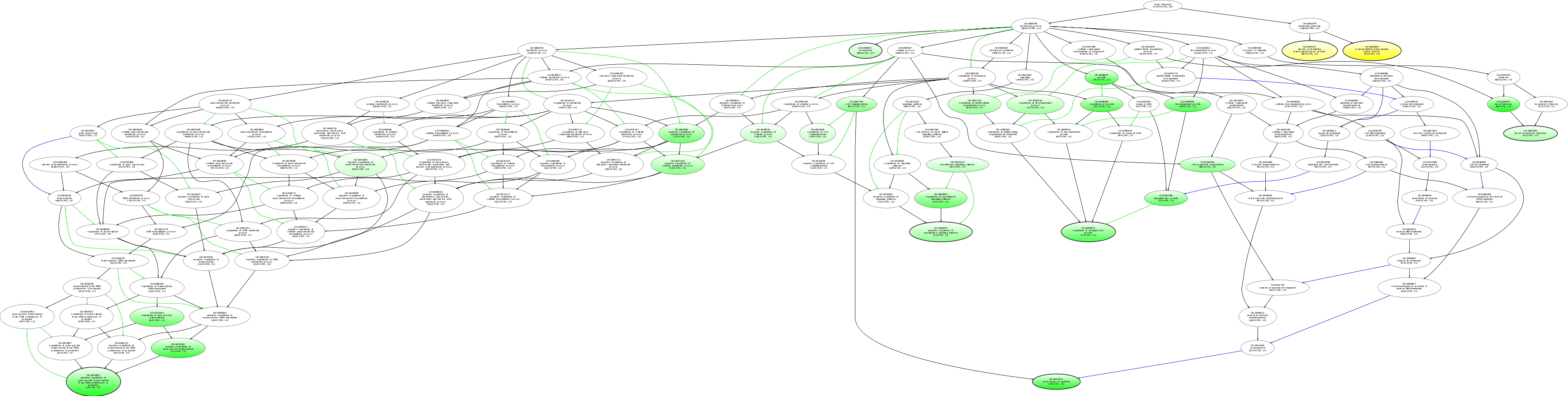
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0000988
|
protein binding transcription factor activity
|
0.0122549 |
0.0122549 |
1
|
68
|
|
GO:0030537
|
larval behavior
|
0.0128542 |
0.0128542 |
1
|
44
|
|
GO:0007412
|
axon target recognition
|
0.0148148 |
0.0148148 |
1
|
10
|
|
GO:0010553
|
negative regulation of gene-specific transcription from RNA polymerase II promoter
|
0.0361446 |
0.0361446 |
1
|
3
|
|
GO:0040007
|
growth
|
0.0459943 |
0.0459943 |
1
|
247
|
|
GO:0032582
|
negative regulation of gene-specific transcription
|
0.0491071 |
0.0491071 |
1
|
11
|
|
GO:0040008
|
regulation of growth
|
0.0574413 |
0.0574413 |
1
|
132
|
|
GO:0048589
|
developmental growth
|
0.0576779 |
0.0576779 |
1
|
154
|
|
GO:0045570
|
regulation of imaginal disc growth
|
0.0590278 |
0.0590278 |
1
|
17
|
|
GO:0007446
|
imaginal disc growth
|
0.0663082 |
0.0663082 |
1
|
37
|
|
GO:0008589
|
regulation of smoothened signaling pathway
|
0.0674157 |
0.0674157 |
1
|
24
|
|
GO:0032583
|
regulation of gene-specific transcription
|
0.0677419 |
0.0677419 |
1
|
42
|
|
GO:0009892
|
negative regulation of metabolic process
|
0.0691964 |
0.0691964 |
1
|
372
|
|
GO:0001071
|
nucleic acid binding transcription factor activity
|
0.0701313 |
0.0701313 |
1
|
395
|
|
GO:0007154
|
cell communication
|
0.0702744 |
0.0702744 |
1
|
461
|
|
GO:0030030
|
cell projection organization
|
0.0738541 |
0.0738541 |
1
|
485
|
|
GO:0031324
|
negative regulation of cellular metabolic process
|
0.0793138 |
0.0793138 |
1
|
319
|
|
GO:0045879
|
negative regulation of smoothened signaling pathway
|
0.0802469 |
0.0802469 |
1
|
13
|
|
GO:0051239
|
regulation of multicellular organismal process
|
0.0844424 |
0.0844424 |
1
|
368
|
|
GO:0050793
|
regulation of developmental process
|
0.0844959 |
0.0844959 |
1
|
321
|
|
GO:0008345
|
larval locomotory behavior
|
0.0874317 |
0.0874317 |
1
|
16
|
|
GO:0040011
|
locomotion
|
0.0891086 |
0.0891086 |
1
|
484
|
|
GO:0007224
|
smoothened signaling pathway
|
0.0929487 |
0.0929487 |
1
|
58
|
|
GO:0048523
|
negative regulation of cellular process
|
0.0966514 |
0.0966514 |
1
|
635
|
|
GO:0010646
|
regulation of cell communication
|
0.0981201 |
0.0981201 |
1
|
214
|
|
GO:0010605
|
negative regulation of macromolecule metabolic process
|
0.0988340 |
0.0988340 |
1
|
356
|
|
GO:0032569
|
gene-specific transcription from RNA polymerase II promoter
|
0.102767 |
0.102767 |
1
|
26
|
|
GO:2000026
|
regulation of multicellular organismal development
|
0.109792 |
0.109792 |
1
|
259
|
|
GO:0048736
|
appendage development
|
0.110896 |
0.110896 |
1
|
286
|
|
GO:0051172
|
negative regulation of nitrogen compound metabolic process
|
0.114943 |
0.114943 |
1
|
250
|
|
GO:0048638
|
regulation of developmental growth
|
0.118943 |
0.118943 |
1
|
54
|
|
GO:0035467
|
negative regulation of signaling pathway
|
0.122789 |
0.122789 |
1
|
118
|
|
GO:0035466
|
regulation of signaling pathway
|
0.126661 |
0.126661 |
1
|
324
|
|
GO:0048519
|
negative regulation of biological process
|
0.128590 |
0.128590 |
1
|
706
|
|
GO:0009890
|
negative regulation of biosynthetic process
|
0.129715 |
0.129715 |
1
|
282
|
|
GO:0010551
|
regulation of gene-specific transcription from RNA polymerase II promoter
|
0.130000 |
0.130000 |
1
|
26
|
|
GO:0007447
|
imaginal disc pattern formation
|
0.133075 |
0.133075 |
1
|
103
|
|
GO:0045934
|
negative regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.135355 |
0.135355 |
1
|
250
|
|
GO:0031327
|
negative regulation of cellular biosynthetic process
|
0.136166 |
0.136166 |
1
|
282
|
|
GO:0010648
|
negative regulation of cell communication
|
0.136511 |
0.136511 |
1
|
126
|
|
GO:0000122
|
negative regulation of transcription from RNA polymerase II promoter
|
0.145455 |
0.145455 |
1
|
56
|
|
GO:0042461
|
photoreceptor cell development
|
0.147601 |
0.147601 |
1
|
80
|
|
GO:0010629
|
negative regulation of gene expression
|
0.149146 |
0.149146 |
1
|
288
|
|
GO:0051253
|
negative regulation of RNA metabolic process
|
0.155744 |
0.155744 |
1
|
202
|
|
GO:0010558
|
negative regulation of macromolecule biosynthetic process
|
0.165094 |
0.165094 |
1
|
280
|
|
GO:2000113
|
negative regulation of cellular macromolecule biosynthetic process
|
0.172946 |
0.172946 |
1
|
280
|
|
GO:0048869
|
cellular developmental process
|
0.176780 |
0.176780 |
1
|
1276
|
|
GO:0035107
|
appendage morphogenesis
|
0.184365 |
0.184365 |
1
|
283
|
|
GO:0009791
|
post-embryonic development
|
0.189538 |
0.189538 |
1
|
500
|
|
GO:0032989
|
cellular component morphogenesis
|
0.193108 |
0.193108 |
1
|
566
|
|
GO:0007389
|
pattern specification process
|
0.203563 |
0.203563 |
1
|
537
|
|
GO:0019222
|
regulation of metabolic process
|
0.210401 |
0.210401 |
1
|
1242
|
|
GO:0035222
|
wing disc pattern formation
|
0.212389 |
0.212389 |
1
|
72
|
|
GO:0035265
|
organ growth
|
0.214575 |
0.214575 |
1
|
53
|
|
GO:0046620
|
regulation of organ growth
|
0.229730 |
0.229730 |
1
|
34
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.237215 |
0.237215 |
1
|
1039
|
|
GO:0023052
|
signaling
|
0.240473 |
0.240473 |
1
|
1364
|
|
GO:0009605
|
response to external stimulus
|
0.242248 |
0.242248 |
1
|
375
|
|
GO:0016481
|
negative regulation of transcription
|
0.243724 |
0.243724 |
1
|
233
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.244709 |
0.244709 |
1
|
1110
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.259446 |
0.259446 |
1
|
412
|
|
GO:0043167
|
ion binding
|
0.265556 |
0.265556 |
1
|
1498
|
|
GO:0045892
|
negative regulation of transcription, DNA-dependent
|
0.266030 |
0.266030 |
1
|
195
|
|
GO:0050896
|
response to stimulus
|
0.270384 |
0.270384 |
1
|
1548
|
|
GO:0007552
|
metamorphosis
|
0.273794 |
0.273794 |
1
|
420
|
|
GO:0048569
|
post-embryonic organ development
|
0.276167 |
0.276167 |
1
|
343
|
|
GO:0007626
|
locomotory behavior
|
0.280859 |
0.280859 |
1
|
157
|
|
GO:0006357
|
regulation of transcription from RNA polymerase II promoter
|
0.282934 |
0.282934 |
1
|
189
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.283034 |
0.283034 |
1
|
1056
|
|
GO:0009953
|
dorsal/ventral pattern formation
|
0.286561 |
0.286561 |
1
|
145
|
|
GO:0006935
|
chemotaxis
|
0.287630 |
0.287630 |
1
|
193
|
|
GO:0032774
|
RNA biosynthetic process
|
0.290017 |
0.290017 |
1
|
703
|
|
GO:0046530
|
photoreceptor cell differentiation
|
0.295620 |
0.295620 |
1
|
162
|
|
GO:0007450
|
dorsal/ventral pattern formation, imaginal disc
|
0.296296 |
0.296296 |
1
|
56
|
|
GO:0050794
|
regulation of cellular process
|
0.306371 |
0.306371 |
1
|
2034
|
|
GO:0003714
|
transcription corepressor activity
|
0.338235 |
0.338235 |
1
|
23
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.357722 |
0.357722 |
1
|
2108
|
|
GO:0006366
|
transcription from RNA polymerase II promoter
|
0.360913 |
0.360913 |
1
|
253
|
|
GO:0007610
|
behavior
|
0.361111 |
0.361111 |
1
|
559
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.366791 |
0.366791 |
1
|
888
|
|
GO:0009887
|
organ morphogenesis
|
0.374384 |
0.374384 |
1
|
684
|
|
GO:0007444
|
imaginal disc development
|
0.376006 |
0.376006 |
1
|
467
|
|
GO:0000904
|
cell morphogenesis involved in differentiation
|
0.376731 |
0.376731 |
1
|
408
|
|
GO:0050789
|
regulation of biological process
|
0.378390 |
0.378390 |
1
|
2246
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.381695 |
0.381695 |
1
|
905
|
|
GO:0006350
|
transcription
|
0.387810 |
0.387810 |
1
|
859
|
|
GO:0042330
|
taxis
|
0.389803 |
0.389803 |
1
|
237
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.396252 |
0.396252 |
1
|
888
|
|
GO:0009058
|
biosynthetic process
|
0.402387 |
0.402387 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.402965 |
0.402965 |
1
|
2093
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.405649 |
0.405649 |
1
|
1623
|
|
GO:0042221
|
response to chemical stimulus
|
0.408269 |
0.408269 |
1
|
632
|
|
GO:0016070
|
RNA metabolic process
|
0.413829 |
0.413829 |
1
|
1191
|
|
GO:0048563
|
post-embryonic organ morphogenesis
|
0.415167 |
0.415167 |
1
|
323
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.415756 |
0.415756 |
1
|
1789
|
|
GO:0065007
|
biological regulation
|
0.422440 |
0.422440 |
1
|
2548
|
|
GO:0048666
|
neuron development
|
0.433702 |
0.433702 |
1
|
471
|
|
GO:0032502
|
developmental process
|
0.435255 |
0.435255 |
1
|
2638
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.440308 |
0.440308 |
1
|
686
|
|
GO:0030182
|
neuron differentiation
|
0.442292 |
0.442292 |
1
|
548
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.451726 |
0.451726 |
1
|
903
|
|
GO:0022008
|
neurogenesis
|
0.452398 |
0.452398 |
1
|
632
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.456989 |
0.456989 |
1
|
850
|
|
GO:0048468
|
cell development
|
0.457419 |
0.457419 |
1
|
1042
|
|
GO:0005575
|
cellular_component
|
0.478421 |
0.478421 |
2
|
8817
|
|
GO:0010468
|
regulation of gene expression
|
0.489930 |
0.489930 |
1
|
973
|
|
GO:0044237
|
cellular metabolic process
|
0.490989 |
0.490989 |
1
|
3814
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.496927 |
0.496927 |
1
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.498346 |
0.498346 |
1
|
1959
|
|
GO:0007399
|
nervous system development
|
0.500000 |
0.500000 |
1
|
848
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.513595 |
0.513595 |
1
|
850
|
|
GO:0044249
|
cellular biosynthetic process
|
0.526807 |
0.526807 |
1
|
2034
|
|
GO:0032501
|
multicellular organismal process
|
0.527040 |
0.527040 |
1
|
3315
|
|
GO:0010467
|
gene expression
|
0.531494 |
0.531494 |
1
|
1907
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0007560
|
imaginal disc morphogenesis
|
0.575758 |
0.575758 |
1
|
323
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0048190
|
wing disc dorsal/ventral pattern formation
|
0.636364 |
0.636364 |
1
|
49
|
|
GO:0035120
|
post-embryonic appendage morphogenesis
|
0.638095 |
0.638095 |
1
|
268
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.639129 |
0.639129 |
1
|
2878
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0043226
|
organelle
|
0.641414 |
0.641414 |
1
|
3537
|
|
GO:0007166
|
cell surface receptor linked signaling pathway
|
0.652720 |
0.652720 |
1
|
624
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0007472
|
wing disc morphogenesis
|
0.665796 |
0.665796 |
1
|
255
|
|
GO:0007409
|
axonogenesis
|
0.679389 |
0.679389 |
1
|
267
|
|
GO:0035220
|
wing disc development
|
0.680942 |
0.680942 |
1
|
318
|
|
GO:0007411
|
axon guidance
|
0.683824 |
0.683824 |
1
|
186
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.684327 |
0.684327 |
1
|
620
|
|
GO:0045449
|
regulation of transcription
|
0.687166 |
0.687166 |
1
|
771
|
|
GO:0043170
|
macromolecule metabolic process
|
0.690797 |
0.690797 |
1
|
3588
|
|
GO:0008150
|
biological_process
|
0.693584 |
0.693584 |
2
|
10616
|
|
GO:0031175
|
neuron projection development
|
0.696270 |
0.696270 |
1
|
392
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0044424
|
intracellular part
|
0.707667 |
0.707667 |
1
|
4384
|
|
GO:0008152
|
metabolic process
|
0.739170 |
0.739170 |
1
|
5194
|
|
GO:0032990
|
cell part morphogenesis
|
0.745583 |
0.745583 |
1
|
422
|
|
GO:0003674
|
molecular_function
|
0.753361 |
0.753361 |
2
|
11064
|
|
GO:0005488
|
binding
|
0.759777 |
0.759777 |
1
|
5641
|
|
GO:0005622
|
intracellular
|
0.764165 |
0.764165 |
1
|
4734
|
|
GO:0008270
|
zinc ion binding
|
0.781440 |
0.781440 |
1
|
901
|
|
GO:0048667
|
cell morphogenesis involved in neuron differentiation
|
0.795501 |
0.795501 |
1
|
389
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0043229
|
intracellular organelle
|
0.804100 |
0.804100 |
1
|
3530
|
|
GO:0048858
|
cell projection morphogenesis
|
0.806883 |
0.806883 |
1
|
422
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.814170 |
0.814170 |
1
|
701
|
|
GO:0044238
|
primary metabolic process
|
0.819985 |
0.819985 |
1
|
4259
|
|
GO:0000902
|
cell morphogenesis
|
0.825088 |
0.825088 |
1
|
467
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.842767 |
0.842767 |
1
|
402
|
|
GO:0009987
|
cellular process
|
0.854049 |
0.854049 |
1
|
6560
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0007476
|
imaginal disc-derived wing morphogenesis
|
0.903915 |
0.903915 |
1
|
254
|
|
GO:0048812
|
neuron projection morphogenesis
|
0.910798 |
0.910798 |
1
|
388
|
|
GO:0005623
|
cell
|
0.911589 |
0.911589 |
1
|
6195
|
|
GO:0044464
|
cell part
|
0.911589 |
0.911589 |
1
|
6195
|
|
GO:0003002
|
regionalization
|
0.942272 |
0.942272 |
1
|
506
|
|
GO:0002165
|
instar larval or pupal development
|
0.954000 |
0.954000 |
1
|
477
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0035114
|
imaginal disc-derived appendage morphogenesis
|
0.975524 |
0.975524 |
1
|
279
|
|
GO:0048737
|
imaginal disc-derived appendage development
|
0.986014 |
0.986014 |
1
|
282
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
2
|
12747
|
|
GO:0000989
|
transcription factor binding transcription factor activity
|
1.00000 |
1.00000 |
1
|
68
|
|
GO:0003712
|
transcription cofactor activity
|
1.00000 |
1.00000 |
1
|
68
|
|
GO:0003700
|
sequence-specific DNA binding transcription factor activity
|
1.00000 |
1.00000 |
1
|
395
|