| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
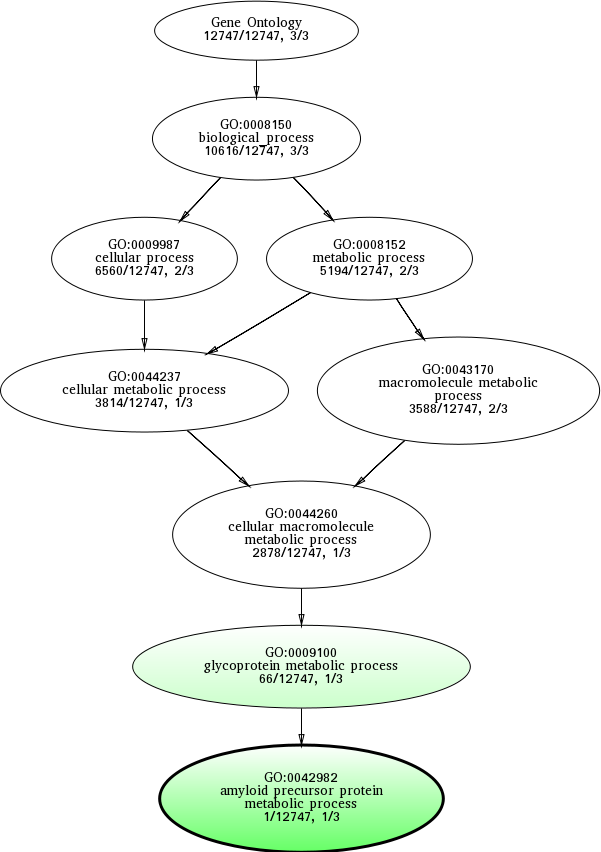
GO:0042982
|
amyloid precursor protein metabolic process
|
0.0151515 |
0.0151515 |
1
|
1
|
|
GO:0009100
|
glycoprotein metabolic process
|
0.0229326 |
0.0229326 |
1
|
66
|
|
GO:0003824
|
catalytic activity
|
0.137704 |
0.137704 |
2
|
4106
|
|
GO:0004222
|
metalloendopeptidase activity
|
0.151460 |
0.151460 |
1
|
83
|
|
GO:0045184
|
establishment of protein localization
|
0.168623 |
0.168623 |
1
|
273
|
|
GO:0015031
|
protein transport
|
0.177333 |
0.177333 |
1
|
266
|
|
GO:0019001
|
guanyl nucleotide binding
|
0.182875 |
0.182875 |
1
|
173
|
|
GO:0032561
|
guanyl ribonucleotide binding
|
0.194098 |
0.194098 |
1
|
171
|
|
GO:0023046
|
signaling process
|
0.199779 |
0.199779 |
1
|
760
|
|
GO:0000166
|
nucleotide binding
|
0.208828 |
0.208828 |
1
|
1178
|
|
GO:0044425
|
membrane part
|
0.225666 |
0.225666 |
1
|
1398
|
|
GO:0016787
|
hydrolase activity
|
0.230601 |
0.230601 |
2
|
1972
|
|
GO:0003924
|
GTPase activity
|
0.241796 |
0.241796 |
1
|
140
|
|
GO:0007165
|
signal transduction
|
0.254057 |
0.254057 |
1
|
548
|
|
GO:0008237
|
metallopeptidase activity
|
0.269481 |
0.269481 |
1
|
166
|
|
GO:0033036
|
macromolecule localization
|
0.276371 |
0.276371 |
1
|
524
|
|
GO:0006508
|
proteolysis
|
0.335119 |
0.335119 |
1
|
688
|
|
GO:0023052
|
signaling
|
0.338079 |
0.338079 |
1
|
1364
|
|
GO:0016020
|
membrane
|
0.342534 |
0.342534 |
1
|
2122
|
|
GO:0051234
|
establishment of localization
|
0.377001 |
0.377001 |
1
|
1549
|
|
GO:0023034
|
intracellular signaling pathway
|
0.430962 |
0.430962 |
1
|
412
|
|
GO:0051179
|
localization
|
0.445834 |
0.445834 |
1
|
1896
|
|
GO:0043170
|
macromolecule metabolic process
|
0.477159 |
0.477159 |
2
|
3588
|
|
GO:0008152
|
metabolic process
|
0.483893 |
0.483893 |
2
|
5194
|
|
GO:0035556
|
intracellular signal transduction
|
0.499208 |
0.499208 |
1
|
315
|
|
GO:0050789
|
regulation of biological process
|
0.509927 |
0.509927 |
1
|
2246
|
|
GO:0016817
|
hydrolase activity, acting on acid anhydrides
|
0.511809 |
0.511809 |
1
|
594
|
|
GO:0050794
|
regulation of cellular process
|
0.518911 |
0.518911 |
1
|
2034
|
|
GO:0008233
|
peptidase activity
|
0.542499 |
0.542499 |
1
|
638
|
|
GO:0065007
|
biological regulation
|
0.561089 |
0.561089 |
1
|
2548
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0007264
|
small GTPase mediated signal transduction
|
0.587302 |
0.587302 |
1
|
185
|
|
GO:0009987
|
cellular process
|
0.673638 |
0.673638 |
2
|
6560
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0005623
|
cell
|
0.702620 |
0.702620 |
1
|
6195
|
|
GO:0044464
|
cell part
|
0.702620 |
0.702620 |
1
|
6195
|
|
GO:0019538
|
protein metabolic process
|
0.724907 |
0.724907 |
1
|
2053
|
|
GO:0031224
|
intrinsic to membrane
|
0.725322 |
0.725322 |
1
|
1014
|
|
GO:0008104
|
protein localization
|
0.727099 |
0.727099 |
1
|
381
|
|
GO:0032553
|
ribonucleotide binding
|
0.746180 |
0.746180 |
1
|
879
|
|
GO:0004175
|
endopeptidase activity
|
0.754870 |
0.754870 |
1
|
465
|
|
GO:0005488
|
binding
|
0.759777 |
0.759777 |
1
|
5641
|
|
GO:0017076
|
purine nucleotide binding
|
0.803056 |
0.803056 |
1
|
946
|
|
GO:0044237
|
cellular metabolic process
|
0.868168 |
0.868168 |
1
|
3814
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.869824 |
0.869824 |
1
|
2878
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.929175 |
0.929175 |
1
|
879
|
|
GO:0003674
|
molecular_function
|
0.952326 |
0.952326 |
2
|
11064
|
|
GO:0070011
|
peptidase activity, acting on L-amino acid peptides
|
0.965517 |
0.965517 |
1
|
616
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0044238
|
primary metabolic process
|
0.967623 |
0.967623 |
1
|
4259
|
|
GO:0005575
|
cellular_component
|
0.970710 |
0.970710 |
1
|
8817
|
|
GO:0017111
|
nucleoside-triphosphatase activity
|
0.983022 |
0.983022 |
1
|
579
|
|
GO:0016021
|
integral to membrane
|
0.984221 |
0.984221 |
1
|
998
|
|
GO:0016462
|
pyrophosphatase activity
|
0.991582 |
0.991582 |
1
|
589
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0016818
|
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
|
1.00000 |
1.00000 |
1
|
594
|
|
GO:0005525
|
GTP binding
|
1.00000 |
1.00000 |
1
|
171
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|