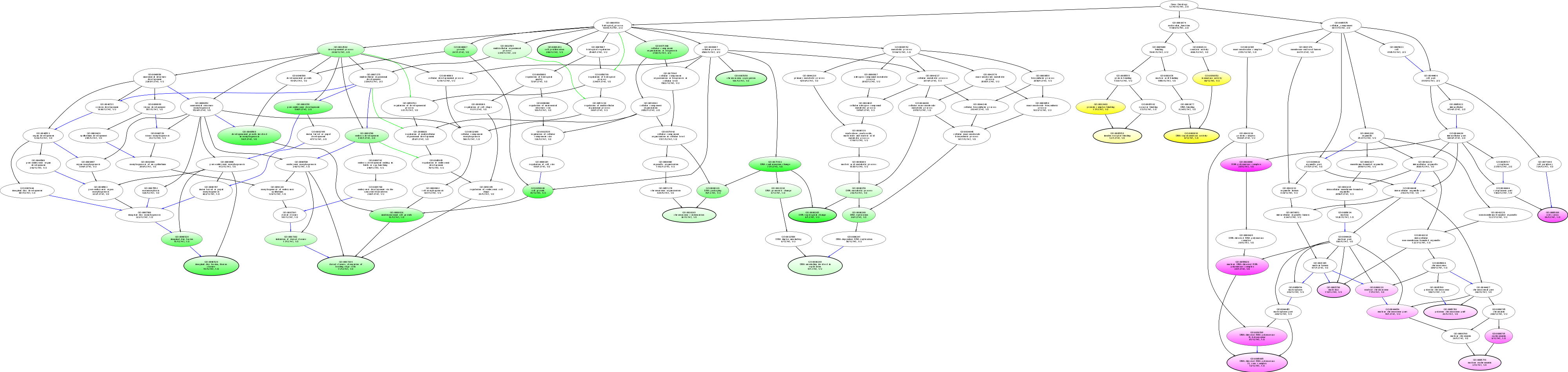
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0003916
|
DNA topoisomerase activity
|
0.00874317 |
0.00874317 |
1
|
8
|
|
GO:0032403
|
protein complex binding
|
0.00984936 |
0.00984936 |
1
|
17
|
|
GO:0030880
|
RNA polymerase complex
|
0.0122150 |
0.0122150 |
1
|
28
|
|
GO:0006265
|
DNA topological change
|
0.0204082 |
0.0204082 |
1
|
6
|
|
GO:0071103
|
DNA conformation change
|
0.0233396 |
0.0233396 |
1
|
77
|
|
GO:0016853
|
isomerase activity
|
0.0241111 |
0.0241111 |
1
|
99
|
|
GO:0016049
|
cell growth
|
0.0274662 |
0.0274662 |
1
|
91
|
|
GO:0009826
|
unidimensional cell growth
|
0.0308285 |
0.0308285 |
1
|
16
|
|
GO:0055029
|
nuclear DNA-directed RNA polymerase complex
|
0.0337349 |
0.0337349 |
1
|
28
|
|
GO:0060560
|
developmental growth involved in morphogenesis
|
0.0355105 |
0.0355105 |
1
|
56
|
|
GO:0009791
|
post-embryonic development
|
0.0358662 |
0.0358662 |
2
|
500
|
|
GO:0046529
|
imaginal disc fusion, thorax closure
|
0.0364078 |
0.0364078 |
1
|
15
|
|
GO:0005938
|
cell cortex
|
0.0369141 |
0.0369141 |
1
|
89
|
|
GO:0008283
|
cell proliferation
|
0.0373258 |
0.0373258 |
1
|
200
|
|
GO:0016591
|
DNA-directed RNA polymerase II, holoenzyme
|
0.0378177 |
0.0378177 |
1
|
61
|
|
GO:0046528
|
imaginal disc fusion
|
0.0388350 |
0.0388350 |
1
|
16
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.0394143 |
0.0394143 |
2
|
2108
|
|
GO:0007394
|
dorsal closure, elongation of leading edge cells
|
0.0431373 |
0.0431373 |
1
|
11
|
|
GO:0000791
|
euchromatin
|
0.0450000 |
0.0450000 |
1
|
9
|
|
GO:0040007
|
growth
|
0.0459943 |
0.0459943 |
1
|
247
|
|
GO:0006323
|
DNA packaging
|
0.0460224 |
0.0460224 |
1
|
70
|
|
GO:0005665
|
DNA-directed RNA polymerase II, core complex
|
0.0490566 |
0.0490566 |
1
|
13
|
|
GO:0007059
|
chromosome segregation
|
0.0499732 |
0.0499732 |
1
|
166
|
|
GO:0006260
|
DNA replication
|
0.0518433 |
0.0518433 |
1
|
90
|
|
GO:0009790
|
embryo development
|
0.0589730 |
0.0589730 |
2
|
641
|
|
GO:0032502
|
developmental process
|
0.0617311 |
0.0617311 |
2
|
2638
|
|
GO:0005730
|
nucleolus
|
0.0667044 |
0.0667044 |
1
|
118
|
|
GO:0044454
|
nuclear chromosome part
|
0.0667939 |
0.0667939 |
1
|
70
|
|
GO:0005158
|
insulin receptor binding
|
0.0681818 |
0.0681818 |
1
|
12
|
|
GO:0007392
|
initiation of dorsal closure
|
0.0699588 |
0.0699588 |
1
|
17
|
|
GO:0000228
|
nuclear chromosome
|
0.0714949 |
0.0714949 |
1
|
77
|
|
GO:0005719
|
nuclear euchromatin
|
0.0731707 |
0.0731707 |
1
|
3
|
|
GO:0005703
|
polytene chromosome puff
|
0.0773333 |
0.0773333 |
1
|
29
|
|
GO:0032392
|
DNA geometric change
|
0.0779221 |
0.0779221 |
1
|
6
|
|
GO:0006259
|
DNA metabolic process
|
0.0840862 |
0.0840862 |
1
|
242
|
|
GO:0030261
|
chromosome condensation
|
0.0849057 |
0.0849057 |
1
|
36
|
|
GO:0006268
|
DNA unwinding involved in replication
|
0.0877193 |
0.0877193 |
1
|
5
|
|
GO:0032501
|
multicellular organismal process
|
0.0974889 |
0.0974889 |
2
|
3315
|
|
GO:0005102
|
receptor binding
|
0.100232 |
0.100232 |
1
|
173
|
|
GO:0051716
|
cellular response to stimulus
|
0.102810 |
0.102810 |
1
|
374
|
|
GO:0048589
|
developmental growth
|
0.112049 |
0.112049 |
1
|
154
|
|
GO:0032535
|
regulation of cellular component size
|
0.122375 |
0.122375 |
1
|
130
|
|
GO:0023014
|
signal transmission via phosphorylation event
|
0.126316 |
0.126316 |
1
|
96
|
|
GO:0023046
|
signaling process
|
0.138061 |
0.138061 |
1
|
760
|
|
GO:0031974
|
membrane-enclosed lumen
|
0.141385 |
0.141385 |
1
|
647
|
|
GO:0008360
|
regulation of cell shape
|
0.153635 |
0.153635 |
1
|
112
|
|
GO:0000790
|
nuclear chromatin
|
0.155172 |
0.155172 |
1
|
36
|
|
GO:0044427
|
chromosomal part
|
0.156236 |
0.156236 |
1
|
342
|
|
GO:0051239
|
regulation of multicellular organismal process
|
0.161772 |
0.161772 |
1
|
368
|
|
GO:0050793
|
regulation of developmental process
|
0.161873 |
0.161873 |
1
|
321
|
|
GO:0031098
|
stress-activated protein kinase signaling cascade
|
0.170886 |
0.170886 |
1
|
54
|
|
GO:0045995
|
regulation of embryonic development
|
0.174308 |
0.174308 |
1
|
70
|
|
GO:0000003
|
reproduction
|
0.183280 |
0.183280 |
1
|
1022
|
|
GO:0022414
|
reproductive process
|
0.183280 |
0.183280 |
1
|
1022
|
|
GO:0035107
|
appendage morphogenesis
|
0.184365 |
0.184365 |
1
|
283
|
|
GO:0016476
|
regulation of embryonic cell shape
|
0.192053 |
0.192053 |
1
|
29
|
|
GO:0048729
|
tissue morphogenesis
|
0.194151 |
0.194151 |
1
|
312
|
|
GO:2000026
|
regulation of multicellular organismal development
|
0.207572 |
0.207572 |
1
|
259
|
|
GO:0002168
|
instar larval development
|
0.208489 |
0.208489 |
1
|
55
|
|
GO:0048736
|
appendage development
|
0.209532 |
0.209532 |
1
|
286
|
|
GO:0071944
|
cell periphery
|
0.220095 |
0.220095 |
1
|
724
|
|
GO:0005875
|
microtubule associated complex
|
0.224768 |
0.224768 |
1
|
363
|
|
GO:0044430
|
cytoskeletal part
|
0.231674 |
0.231674 |
1
|
512
|
|
GO:0090066
|
regulation of anatomical structure size
|
0.231824 |
0.231824 |
1
|
169
|
|
GO:0023052
|
signaling
|
0.240473 |
0.240473 |
1
|
1364
|
|
GO:0009888
|
tissue development
|
0.245916 |
0.245916 |
1
|
557
|
|
GO:0007165
|
signal transduction
|
0.254057 |
0.254057 |
1
|
548
|
|
GO:0048598
|
embryonic morphogenesis
|
0.255261 |
0.255261 |
1
|
243
|
|
GO:0005488
|
binding
|
0.259926 |
0.259926 |
2
|
5641
|
|
GO:0050896
|
response to stimulus
|
0.270384 |
0.270384 |
1
|
1548
|
|
GO:0044428
|
nuclear part
|
0.271419 |
0.271419 |
1
|
830
|
|
GO:0007552
|
metamorphosis
|
0.273794 |
0.273794 |
1
|
420
|
|
GO:0048569
|
post-embryonic organ development
|
0.276167 |
0.276167 |
1
|
343
|
|
GO:0005700
|
polytene chromosome
|
0.276650 |
0.276650 |
1
|
109
|
|
GO:0065008
|
regulation of biological quality
|
0.286107 |
0.286107 |
1
|
729
|
|
GO:0043233
|
organelle lumen
|
0.291360 |
0.291360 |
1
|
634
|
|
GO:0070013
|
intracellular organelle lumen
|
0.293247 |
0.293247 |
1
|
634
|
|
GO:0005737
|
cytoplasm
|
0.294170 |
0.294170 |
2
|
2378
|
|
GO:0007243
|
intracellular protein kinase cascade
|
0.304762 |
0.304762 |
1
|
96
|
|
GO:0002164
|
larval development
|
0.311383 |
0.311383 |
1
|
85
|
|
GO:0044451
|
nucleoplasm part
|
0.313253 |
0.313253 |
1
|
260
|
|
GO:0005694
|
chromosome
|
0.321108 |
0.321108 |
1
|
394
|
|
GO:0048869
|
cellular developmental process
|
0.322329 |
0.322329 |
1
|
1276
|
|
GO:0005654
|
nucleoplasm
|
0.340964 |
0.340964 |
1
|
283
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.346904 |
0.346904 |
1
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.347592 |
0.347592 |
1
|
1227
|
|
GO:0016331
|
morphogenesis of embryonic epithelium
|
0.347709 |
0.347709 |
1
|
129
|
|
GO:0032989
|
cellular component morphogenesis
|
0.348979 |
0.348979 |
1
|
566
|
|
GO:0051276
|
chromosome organization
|
0.358714 |
0.358714 |
1
|
424
|
|
GO:0009887
|
organ morphogenesis
|
0.374384 |
0.374384 |
1
|
684
|
|
GO:0003918
|
DNA topoisomerase (ATP-hydrolyzing) activity
|
0.375000 |
0.375000 |
1
|
3
|
|
GO:0007444
|
imaginal disc development
|
0.376006 |
0.376006 |
1
|
467
|
|
GO:0050789
|
regulation of biological process
|
0.378390 |
0.378390 |
1
|
2246
|
|
GO:0009987
|
cellular process
|
0.381822 |
0.381822 |
2
|
6560
|
|
GO:0006950
|
response to stress
|
0.390827 |
0.390827 |
1
|
605
|
|
GO:0007391
|
dorsal closure
|
0.392996 |
0.392996 |
1
|
101
|
|
GO:0033554
|
cellular response to stress
|
0.397122 |
0.397122 |
1
|
276
|
|
GO:0009058
|
biosynthetic process
|
0.402387 |
0.402387 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.402965 |
0.402965 |
1
|
2093
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.405649 |
0.405649 |
1
|
1623
|
|
GO:0048563
|
post-embryonic organ morphogenesis
|
0.415167 |
0.415167 |
1
|
323
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.415756 |
0.415756 |
1
|
1789
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.418447 |
0.418447 |
1
|
1562
|
|
GO:0065007
|
biological regulation
|
0.422440 |
0.422440 |
1
|
2548
|
|
GO:0032991
|
macromolecular complex
|
0.428425 |
0.428425 |
1
|
2151
|
|
GO:0023034
|
intracellular signaling pathway
|
0.430962 |
0.430962 |
1
|
412
|
|
GO:0044422
|
organelle part
|
0.432705 |
0.432705 |
1
|
2176
|
|
GO:0007275
|
multicellular organismal development
|
0.442066 |
0.442066 |
2
|
2296
|
|
GO:0003677
|
DNA binding
|
0.444205 |
0.444205 |
1
|
824
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.451701 |
0.451701 |
1
|
412
|
|
GO:0005856
|
cytoskeleton
|
0.457213 |
0.457213 |
1
|
561
|
|
GO:0031981
|
nuclear lumen
|
0.475277 |
0.475277 |
1
|
471
|
|
GO:0005575
|
cellular_component
|
0.478421 |
0.478421 |
2
|
8817
|
|
GO:0048609
|
multicellular organismal reproductive process
|
0.480389 |
0.480389 |
1
|
937
|
|
GO:0032504
|
multicellular organism reproduction
|
0.480389 |
0.480389 |
1
|
937
|
|
GO:0005623
|
cell
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.496927 |
0.496927 |
1
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.498346 |
0.498346 |
1
|
1959
|
|
GO:0035556
|
intracellular signal transduction
|
0.499208 |
0.499208 |
1
|
315
|
|
GO:0003917
|
DNA topoisomerase type I activity
|
0.500000 |
0.500000 |
1
|
4
|
|
GO:0044424
|
intracellular part
|
0.500760 |
0.500760 |
2
|
4384
|
|
GO:0005515
|
protein binding
|
0.518366 |
0.518366 |
1
|
1726
|
|
GO:0050794
|
regulation of cellular process
|
0.518911 |
0.518911 |
1
|
2034
|
|
GO:0044249
|
cellular biosynthetic process
|
0.526807 |
0.526807 |
1
|
2034
|
|
GO:0060429
|
epithelium development
|
0.529623 |
0.529623 |
1
|
295
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0003676
|
nucleic acid binding
|
0.549587 |
0.549587 |
1
|
1855
|
|
GO:0007560
|
imaginal disc morphogenesis
|
0.575758 |
0.575758 |
1
|
323
|
|
GO:0005622
|
intracellular
|
0.583919 |
0.583919 |
2
|
4734
|
|
GO:0000785
|
chromatin
|
0.584795 |
0.584795 |
1
|
200
|
|
GO:0003824
|
catalytic activity
|
0.604523 |
0.604523 |
1
|
4106
|
|
GO:0009792
|
embryo development ending in birth or egg hatching
|
0.609009 |
0.609009 |
1
|
240
|
|
GO:0006261
|
DNA-dependent DNA replication
|
0.622222 |
0.622222 |
1
|
56
|
|
GO:0035120
|
post-embryonic appendage morphogenesis
|
0.638095 |
0.638095 |
1
|
268
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.639129 |
0.639129 |
1
|
2878
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0043226
|
organelle
|
0.641414 |
0.641414 |
1
|
3537
|
|
GO:0007472
|
wing disc morphogenesis
|
0.665796 |
0.665796 |
1
|
255
|
|
GO:0044444
|
cytoplasmic part
|
0.669378 |
0.669378 |
1
|
1863
|
|
GO:0035220
|
wing disc development
|
0.680942 |
0.680942 |
1
|
318
|
|
GO:0043170
|
macromolecule metabolic process
|
0.690797 |
0.690797 |
1
|
3588
|
|
GO:0008150
|
biological_process
|
0.693584 |
0.693584 |
2
|
10616
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0043234
|
protein complex
|
0.727104 |
0.727104 |
1
|
1564
|
|
GO:0007292
|
female gamete generation
|
0.727380 |
0.727380 |
1
|
619
|
|
GO:0008152
|
metabolic process
|
0.739170 |
0.739170 |
1
|
5194
|
|
GO:0044237
|
cellular metabolic process
|
0.740940 |
0.740940 |
1
|
3814
|
|
GO:0044446
|
intracellular organelle part
|
0.743053 |
0.743053 |
1
|
2162
|
|
GO:0003674
|
molecular_function
|
0.753361 |
0.753361 |
2
|
11064
|
|
GO:0008361
|
regulation of cell size
|
0.769231 |
0.769231 |
1
|
100
|
|
GO:0006996
|
organelle organization
|
0.779169 |
0.779169 |
1
|
1182
|
|
GO:0015630
|
microtubule cytoskeleton
|
0.798574 |
0.798574 |
1
|
448
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0044238
|
primary metabolic process
|
0.819985 |
0.819985 |
1
|
4259
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.824951 |
0.824951 |
1
|
1534
|
|
GO:0000902
|
cell morphogenesis
|
0.825088 |
0.825088 |
1
|
467
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0019953
|
sexual reproduction
|
0.859100 |
0.859100 |
1
|
878
|
|
GO:0048731
|
system development
|
0.882863 |
0.882863 |
1
|
1696
|
|
GO:0007276
|
gamete generation
|
0.883697 |
0.883697 |
1
|
851
|
|
GO:0007476
|
imaginal disc-derived wing morphogenesis
|
0.903915 |
0.903915 |
1
|
254
|
|
GO:0002009
|
morphogenesis of an epithelium
|
0.908805 |
0.908805 |
1
|
289
|
|
GO:0002165
|
instar larval or pupal development
|
0.910028 |
0.910028 |
2
|
477
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.923977 |
0.923977 |
1
|
1517
|
|
GO:0016043
|
cellular component organization
|
0.947563 |
0.947563 |
2
|
2052
|
|
GO:0001700
|
embryonic development via the syncytial blastoderm
|
0.958333 |
0.958333 |
1
|
230
|
|
GO:0043229
|
intracellular organelle
|
0.961659 |
0.961659 |
1
|
3530
|
|
GO:0035114
|
imaginal disc-derived appendage morphogenesis
|
0.975524 |
0.975524 |
1
|
279
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.975556 |
0.975556 |
1
|
402
|
|
GO:0048856
|
anatomical structure development
|
0.980053 |
0.980053 |
1
|
2265
|
|
GO:0048737
|
imaginal disc-derived appendage development
|
0.986014 |
0.986014 |
1
|
282
|
|
GO:0048477
|
oogenesis
|
0.987076 |
0.987076 |
1
|
611
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
2
|
12747
|
|
GO:0000428
|
DNA-directed RNA polymerase complex
|
1.00000 |
1.00000 |
1
|
28
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|
|
GO:0032508
|
DNA duplex unwinding
|
1.00000 |
1.00000 |
1
|
6
|