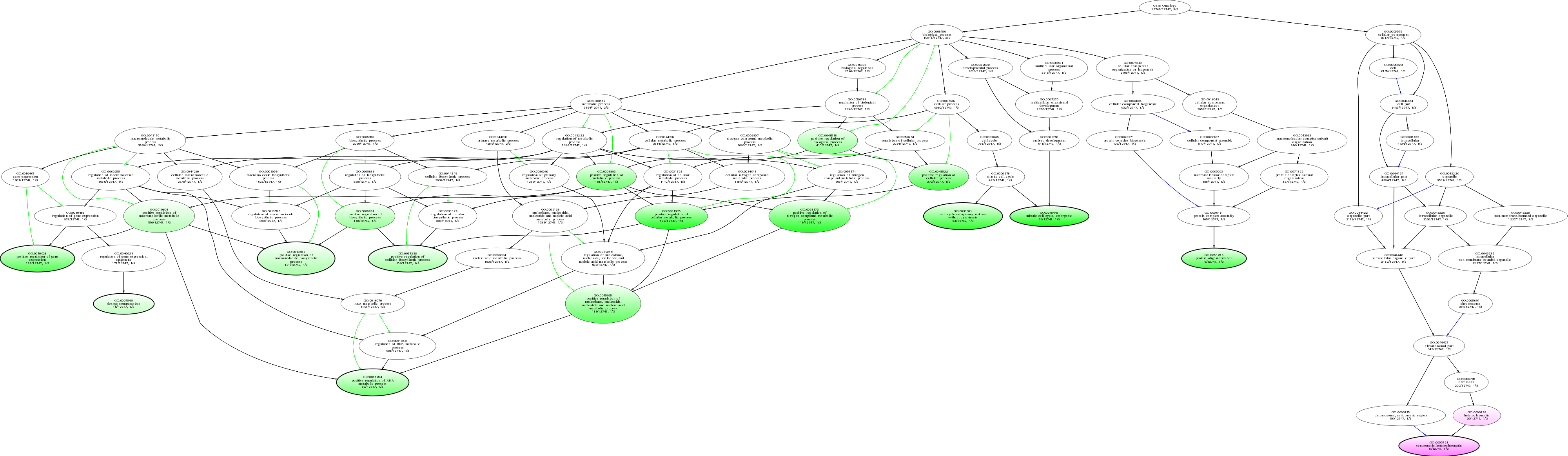
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0045448
|
mitotic cell cycle, embryonic
|
0.0363636 |
0.0363636 |
1
|
36
|
|
GO:0031325
|
positive regulation of cellular metabolic process
|
0.0438776 |
0.0438776 |
1
|
172
|
|
GO:0051173
|
positive regulation of nitrogen compound metabolic process
|
0.0557638 |
0.0557638 |
1
|
119
|
|
GO:0033301
|
cell cycle comprising mitosis without cytokinesis
|
0.0559441 |
0.0559441 |
1
|
24
|
|
GO:0048522
|
positive regulation of cellular process
|
0.0566124 |
0.0566124 |
1
|
372
|
|
GO:0051259
|
protein oligomerization
|
0.0571429 |
0.0571429 |
1
|
6
|
|
GO:0010628
|
positive regulation of gene expression
|
0.0636411 |
0.0636411 |
1
|
122
|
|
GO:0045935
|
positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.0649918 |
0.0649918 |
1
|
119
|
|
GO:0009893
|
positive regulation of metabolic process
|
0.0671169 |
0.0671169 |
1
|
181
|
|
GO:0051254
|
positive regulation of RNA metabolic process
|
0.0676998 |
0.0676998 |
1
|
83
|
|
GO:0005721
|
centromeric heterochromatin
|
0.0746269 |
0.0746269 |
1
|
5
|
|
GO:0048518
|
positive regulation of biological process
|
0.0757538 |
0.0757538 |
1
|
410
|
|
GO:0009891
|
positive regulation of biosynthetic process
|
0.0760818 |
0.0760818 |
1
|
160
|
|
GO:0031328
|
positive regulation of cellular biosynthetic process
|
0.0777886 |
0.0777886 |
1
|
159
|
|
GO:0010557
|
positive regulation of macromolecule biosynthetic process
|
0.0789632 |
0.0789632 |
1
|
131
|
|
GO:0010604
|
positive regulation of macromolecule metabolic process
|
0.0814533 |
0.0814533 |
1
|
150
|
|
GO:0007549
|
dosage compensation
|
0.0903955 |
0.0903955 |
1
|
16
|
|
GO:0000792
|
heterochromatin
|
0.100000 |
0.100000 |
1
|
20
|
|
GO:0048736
|
appendage development
|
0.110896 |
0.110896 |
1
|
286
|
|
GO:0030528
|
transcription regulator activity
|
0.112632 |
0.112632 |
1
|
432
|
|
GO:0045893
|
positive regulation of transcription, DNA-dependent
|
0.114685 |
0.114685 |
1
|
82
|
|
GO:0007049
|
cell cycle
|
0.115244 |
0.115244 |
1
|
756
|
|
GO:0043933
|
macromolecular complex subunit organization
|
0.116959 |
0.116959 |
1
|
240
|
|
GO:0016888
|
endodeoxyribonuclease activity, producing 5'-phosphomonoesters
|
0.121212 |
0.121212 |
1
|
4
|
|
GO:0045941
|
positive regulation of transcription
|
0.127636 |
0.127636 |
1
|
115
|
|
GO:0048285
|
organelle fission
|
0.148900 |
0.148900 |
1
|
176
|
|
GO:0044427
|
chromosomal part
|
0.156236 |
0.156236 |
1
|
342
|
|
GO:0000775
|
chromosome, centromeric region
|
0.163743 |
0.163743 |
1
|
56
|
|
GO:0070271
|
protein complex biogenesis
|
0.166139 |
0.166139 |
1
|
105
|
|
GO:0040029
|
regulation of gene expression, epigenetic
|
0.181912 |
0.181912 |
1
|
177
|
|
GO:0035107
|
appendage morphogenesis
|
0.184365 |
0.184365 |
1
|
283
|
|
GO:0004518
|
nuclease activity
|
0.189516 |
0.189516 |
1
|
94
|
|
GO:0009791
|
post-embryonic development
|
0.189538 |
0.189538 |
1
|
500
|
|
GO:0016787
|
hydrolase activity
|
0.230601 |
0.230601 |
2
|
1972
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.237386 |
0.237386 |
1
|
1562
|
|
GO:0008152
|
metabolic process
|
0.239353 |
0.239353 |
2
|
5194
|
|
GO:0009790
|
embryo development
|
0.242987 |
0.242987 |
1
|
641
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.244709 |
0.244709 |
1
|
1110
|
|
GO:0044422
|
organelle part
|
0.246796 |
0.246796 |
1
|
2176
|
|
GO:0009886
|
post-embryonic morphogenesis
|
0.259446 |
0.259446 |
1
|
412
|
|
GO:0043167
|
ion binding
|
0.265556 |
0.265556 |
1
|
1498
|
|
GO:0007552
|
metamorphosis
|
0.273794 |
0.273794 |
1
|
420
|
|
GO:0022607
|
cellular component assembly
|
0.275418 |
0.275418 |
1
|
577
|
|
GO:0048569
|
post-embryonic organ development
|
0.276167 |
0.276167 |
1
|
343
|
|
GO:0006357
|
regulation of transcription from RNA polymerase II promoter
|
0.282934 |
0.282934 |
1
|
189
|
|
GO:0032774
|
RNA biosynthetic process
|
0.290017 |
0.290017 |
1
|
703
|
|
GO:0003704
|
specific RNA polymerase II transcription factor activity
|
0.292135 |
0.292135 |
1
|
78
|
|
GO:0017171
|
serine hydrolase activity
|
0.294877 |
0.294877 |
1
|
316
|
|
GO:0044085
|
cellular component biogenesis
|
0.299810 |
0.299810 |
1
|
632
|
|
GO:0065003
|
macromolecular complex assembly
|
0.301303 |
0.301303 |
1
|
185
|
|
GO:0005515
|
protein binding
|
0.305974 |
0.305974 |
1
|
1726
|
|
GO:0050794
|
regulation of cellular process
|
0.306371 |
0.306371 |
1
|
2034
|
|
GO:0003824
|
catalytic activity
|
0.310936 |
0.310936 |
2
|
4106
|
|
GO:0005694
|
chromosome
|
0.321108 |
0.321108 |
1
|
394
|
|
GO:0003676
|
nucleic acid binding
|
0.328842 |
0.328842 |
1
|
1855
|
|
GO:0004536
|
deoxyribonuclease activity
|
0.329787 |
0.329787 |
1
|
31
|
|
GO:0006508
|
proteolysis
|
0.335119 |
0.335119 |
1
|
688
|
|
GO:0004520
|
endodeoxyribonuclease activity
|
0.339286 |
0.339286 |
1
|
19
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.346904 |
0.346904 |
1
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.347592 |
0.347592 |
1
|
1227
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.357722 |
0.357722 |
1
|
2108
|
|
GO:0051276
|
chromosome organization
|
0.358714 |
0.358714 |
1
|
424
|
|
GO:0006366
|
transcription from RNA polymerase II promoter
|
0.360913 |
0.360913 |
1
|
253
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.366791 |
0.366791 |
1
|
888
|
|
GO:0009887
|
organ morphogenesis
|
0.374384 |
0.374384 |
1
|
684
|
|
GO:0007444
|
imaginal disc development
|
0.376006 |
0.376006 |
1
|
467
|
|
GO:0019222
|
regulation of metabolic process
|
0.376562 |
0.376562 |
1
|
1242
|
|
GO:0050789
|
regulation of biological process
|
0.378390 |
0.378390 |
1
|
2246
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.381695 |
0.381695 |
1
|
905
|
|
GO:0006350
|
transcription
|
0.387810 |
0.387810 |
1
|
859
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.396252 |
0.396252 |
1
|
888
|
|
GO:0016893
|
endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 5'-phosphomonoesters
|
0.400000 |
0.400000 |
1
|
18
|
|
GO:0043226
|
organelle
|
0.401157 |
0.401157 |
1
|
3537
|
|
GO:0016070
|
RNA metabolic process
|
0.413829 |
0.413829 |
1
|
1191
|
|
GO:0048563
|
post-embryonic organ morphogenesis
|
0.415167 |
0.415167 |
1
|
323
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.418200 |
0.418200 |
1
|
1039
|
|
GO:0065007
|
biological regulation
|
0.422440 |
0.422440 |
1
|
2548
|
|
GO:0032502
|
developmental process
|
0.435255 |
0.435255 |
1
|
2638
|
|
GO:0016788
|
hydrolase activity, acting on ester bonds
|
0.439875 |
0.439875 |
1
|
496
|
|
GO:0051252
|
regulation of RNA metabolic process
|
0.440308 |
0.440308 |
1
|
686
|
|
GO:0003677
|
DNA binding
|
0.444205 |
0.444205 |
1
|
824
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.451726 |
0.451726 |
1
|
903
|
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.456989 |
0.456989 |
1
|
850
|
|
GO:0043170
|
macromolecule metabolic process
|
0.477159 |
0.477159 |
2
|
3588
|
|
GO:0004519
|
endonuclease activity
|
0.478723 |
0.478723 |
1
|
45
|
|
GO:0006461
|
protein complex assembly
|
0.483871 |
0.483871 |
1
|
105
|
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.486014 |
0.486014 |
1
|
1056
|
|
GO:0010468
|
regulation of gene expression
|
0.489930 |
0.489930 |
1
|
973
|
|
GO:0044446
|
intracellular organelle part
|
0.493044 |
0.493044 |
1
|
2162
|
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.496927 |
0.496927 |
1
|
1617
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.498346 |
0.498346 |
1
|
1959
|
|
GO:0008236
|
serine-type peptidase activity
|
0.508091 |
0.508091 |
1
|
314
|
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.513595 |
0.513595 |
1
|
850
|
|
GO:0044249
|
cellular biosynthetic process
|
0.526807 |
0.526807 |
1
|
2034
|
|
GO:0032501
|
multicellular organismal process
|
0.527040 |
0.527040 |
1
|
3315
|
|
GO:0008233
|
peptidase activity
|
0.542499 |
0.542499 |
1
|
638
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0004252
|
serine-type endopeptidase activity
|
0.554890 |
0.554890 |
1
|
278
|
|
GO:0000278
|
mitotic cell cycle
|
0.567460 |
0.567460 |
1
|
429
|
|
GO:0071822
|
protein complex subunit organization
|
0.570833 |
0.570833 |
1
|
137
|
|
GO:0007560
|
imaginal disc morphogenesis
|
0.575758 |
0.575758 |
1
|
323
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0035186
|
syncytial blastoderm mitotic cell cycle
|
0.583333 |
0.583333 |
1
|
21
|
|
GO:0000785
|
chromatin
|
0.584795 |
0.584795 |
1
|
200
|
|
GO:0003702
|
RNA polymerase II transcription factor activity
|
0.618056 |
0.618056 |
1
|
267
|
|
GO:0035120
|
post-embryonic appendage morphogenesis
|
0.638095 |
0.638095 |
1
|
268
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0009058
|
biosynthetic process
|
0.642905 |
0.642905 |
1
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.643595 |
0.643595 |
1
|
2093
|
|
GO:0006325
|
chromatin organization
|
0.643868 |
0.643868 |
1
|
273
|
|
GO:0009059
|
macromolecule biosynthetic process
|
0.646807 |
0.646807 |
1
|
1623
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.658716 |
0.658716 |
1
|
1789
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0007472
|
wing disc morphogenesis
|
0.665796 |
0.665796 |
1
|
255
|
|
GO:0044238
|
primary metabolic process
|
0.672346 |
0.672346 |
2
|
4259
|
|
GO:0035220
|
wing disc development
|
0.680942 |
0.680942 |
1
|
318
|
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
0.684327 |
0.684327 |
1
|
620
|
|
GO:0045449
|
regulation of transcription
|
0.687166 |
0.687166 |
1
|
771
|
|
GO:0005623
|
cell
|
0.702620 |
0.702620 |
1
|
6195
|
|
GO:0044464
|
cell part
|
0.702620 |
0.702620 |
1
|
6195
|
|
GO:0044424
|
intracellular part
|
0.707667 |
0.707667 |
1
|
4384
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.724105 |
0.724105 |
1
|
1517
|
|
GO:0019538
|
protein metabolic process
|
0.724907 |
0.724907 |
1
|
2053
|
|
GO:0044237
|
cellular metabolic process
|
0.740940 |
0.740940 |
1
|
3814
|
|
GO:0004175
|
endopeptidase activity
|
0.754870 |
0.754870 |
1
|
465
|
|
GO:0005622
|
intracellular
|
0.764165 |
0.764165 |
1
|
4734
|
|
GO:0006996
|
organelle organization
|
0.779169 |
0.779169 |
1
|
1182
|
|
GO:0010467
|
gene expression
|
0.780571 |
0.780571 |
1
|
1907
|
|
GO:0008270
|
zinc ion binding
|
0.781440 |
0.781440 |
1
|
901
|
|
GO:0046914
|
transition metal ion binding
|
0.795721 |
0.795721 |
1
|
1153
|
|
GO:0043229
|
intracellular organelle
|
0.804100 |
0.804100 |
1
|
3530
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0006351
|
transcription, DNA-dependent
|
0.814170 |
0.814170 |
1
|
701
|
|
GO:0048707
|
instar larval or pupal morphogenesis
|
0.842767 |
0.842767 |
1
|
402
|
|
GO:0009987
|
cellular process
|
0.854049 |
0.854049 |
1
|
6560
|
|
GO:0090304
|
nucleic acid metabolic process
|
0.858021 |
0.858021 |
1
|
1535
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.869824 |
0.869824 |
1
|
2878
|
|
GO:0005488
|
binding
|
0.882277 |
0.882277 |
1
|
5641
|
|
GO:0007476
|
imaginal disc-derived wing morphogenesis
|
0.903915 |
0.903915 |
1
|
254
|
|
GO:0008150
|
biological_process
|
0.925522 |
0.925522 |
2
|
10616
|
|
GO:0002165
|
instar larval or pupal development
|
0.954000 |
0.954000 |
1
|
477
|
|
GO:0070011
|
peptidase activity, acting on L-amino acid peptides
|
0.965517 |
0.965517 |
1
|
616
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0005575
|
cellular_component
|
0.970710 |
0.970710 |
1
|
8817
|
|
GO:0000280
|
nuclear division
|
0.971591 |
0.971591 |
1
|
171
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0035114
|
imaginal disc-derived appendage morphogenesis
|
0.975524 |
0.975524 |
1
|
279
|
|
GO:0048737
|
imaginal disc-derived appendage development
|
0.986014 |
0.986014 |
1
|
282
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|