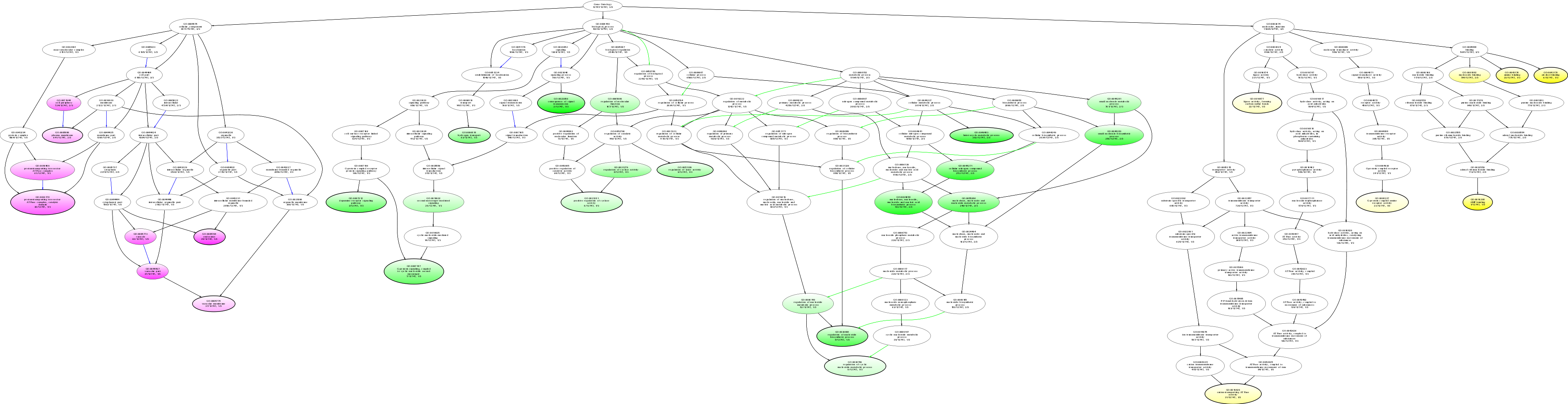
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0023050
|
consequence of signal transmission
|
0.00263158 |
0.00263158 |
1
|
2
|
|
GO:0016208
|
AMP binding
|
0.00280505 |
0.00280505 |
1
|
1
|
|
GO:0043178
|
alcohol binding
|
0.00318810 |
0.00318810 |
1
|
6
|
|
GO:0046483
|
heterocycle metabolic process
|
0.00584290 |
0.00584290 |
2
|
292
|
|
GO:0034654
|
nucleobase, nucleoside, nucleotide and nucleic acid biosynthetic process
|
0.00753916 |
0.00753916 |
2
|
163
|
|
GO:0044271
|
cellular nitrogen compound biosynthetic process
|
0.00865409 |
0.00865409 |
2
|
252
|
|
GO:0005768
|
endosome
|
0.00977654 |
0.00977654 |
1
|
35
|
|
GO:0044283
|
small molecule biosynthetic process
|
0.0110286 |
0.0110286 |
2
|
259
|
|
GO:0044437
|
vacuolar part
|
0.0120718 |
0.0120718 |
1
|
37
|
|
GO:0043176
|
amine binding
|
0.0121842 |
0.0121842 |
1
|
23
|
|
GO:0055086
|
nucleobase, nucleoside and nucleotide metabolic process
|
0.0126263 |
0.0126263 |
2
|
249
|
|
GO:0030808
|
regulation of nucleotide biosynthetic process
|
0.0148904 |
0.0148904 |
1
|
8
|
|
GO:0007212
|
dopamine receptor signaling pathway
|
0.0150754 |
0.0150754 |
1
|
3
|
|
GO:0005773
|
vacuole
|
0.0173184 |
0.0173184 |
1
|
62
|
|
GO:0033178
|
proton-transporting two-sector ATPase complex, catalytic domain
|
0.0236538 |
0.0236538 |
1
|
30
|
|
GO:0007187
|
G-protein signaling, coupled to cyclic nucleotide second messenger
|
0.0343137 |
0.0343137 |
1
|
7
|
|
GO:0071944
|
cell periphery
|
0.0377440 |
0.0377440 |
2
|
724
|
|
GO:0006818
|
hydrogen transport
|
0.0420842 |
0.0420842 |
1
|
63
|
|
GO:0016469
|
proton-transporting two-sector ATPase complex
|
0.0477986 |
0.0477986 |
1
|
61
|
|
GO:0001882
|
nucleoside binding
|
0.0525332 |
0.0525332 |
2
|
784
|
|
GO:0031279
|
regulation of cyclase activity
|
0.0536913 |
0.0536913 |
1
|
8
|
|
GO:0044281
|
small molecule metabolic process
|
0.0580586 |
0.0580586 |
2
|
761
|
|
GO:0051339
|
regulation of lyase activity
|
0.0604027 |
0.0604027 |
1
|
9
|
|
GO:0065009
|
regulation of molecular function
|
0.0655416 |
0.0655416 |
1
|
167
|
|
GO:0019829
|
cation-transporting ATPase activity
|
0.0741627 |
0.0741627 |
1
|
31
|
|
GO:0005886
|
plasma membrane
|
0.0851857 |
0.0851857 |
2
|
641
|
|
GO:0019932
|
second-messenger-mediated signaling
|
0.0920635 |
0.0920635 |
1
|
29
|
|
GO:0005774
|
vacuolar membrane
|
0.0934343 |
0.0934343 |
1
|
37
|
|
GO:0031281
|
positive regulation of cyclase activity
|
0.0937500 |
0.0937500 |
1
|
6
|
|
GO:0006140
|
regulation of nucleotide metabolic process
|
0.0944472 |
0.0944472 |
1
|
52
|
|
GO:0016877
|
ligase activity, forming carbon-sulfur bonds
|
0.0952381 |
0.0952381 |
1
|
22
|
|
GO:0008227
|
G-protein coupled amine receptor activity
|
0.0954357 |
0.0954357 |
1
|
23
|
|
GO:0030799
|
regulation of cyclic nucleotide metabolic process
|
0.0963855 |
0.0963855 |
1
|
8
|
|
GO:0034220
|
ion transmembrane transport
|
0.100159 |
0.100159 |
1
|
63
|
|
GO:0016874
|
ligase activity
|
0.109366 |
0.109366 |
1
|
231
|
|
GO:0051349
|
positive regulation of lyase activity
|
0.109375 |
0.109375 |
1
|
7
|
|
GO:0000166
|
nucleotide binding
|
0.112563 |
0.112563 |
2
|
1178
|
|
GO:0008152
|
metabolic process
|
0.117083 |
0.117083 |
3
|
5194
|
|
GO:0016820
|
hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances
|
0.117697 |
0.117697 |
1
|
139
|
|
GO:0050790
|
regulation of catalytic activity
|
0.119487 |
0.119487 |
1
|
149
|
|
GO:0044425
|
membrane part
|
0.129745 |
0.129745 |
2
|
1398
|
|
GO:0004935
|
adrenergic receptor activity
|
0.130435 |
0.130435 |
1
|
3
|
|
GO:0055085
|
transmembrane transport
|
0.132219 |
0.132219 |
1
|
469
|
|
GO:0005488
|
binding
|
0.132501 |
0.132501 |
3
|
5641
|
|
GO:0003987
|
acetate-CoA ligase activity
|
0.142857 |
0.142857 |
1
|
2
|
|
GO:0060089
|
molecular transducer activity
|
0.144056 |
0.144056 |
1
|
559
|
|
GO:0042625
|
ATPase activity, coupled to transmembrane movement of ions
|
0.150268 |
0.150268 |
1
|
84
|
|
GO:0031090
|
organelle membrane
|
0.153645 |
0.153645 |
1
|
396
|
|
GO:0046058
|
cAMP metabolic process
|
0.154390 |
0.154390 |
1
|
15
|
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.172797 |
0.172797 |
2
|
1789
|
|
GO:0031226
|
intrinsic to plasma membrane
|
0.179931 |
0.179931 |
1
|
208
|
|
GO:0006171
|
cAMP biosynthetic process
|
0.189362 |
0.189362 |
1
|
14
|
|
GO:0023046
|
signaling process
|
0.199779 |
0.199779 |
1
|
760
|
|
GO:0046933
|
hydrogen ion transporting ATP synthase activity, rotational mechanism
|
0.200000 |
0.200000 |
1
|
20
|
|
GO:0005887
|
integral to plasma membrane
|
0.203593 |
0.203593 |
1
|
204
|
|
GO:0035240
|
dopamine binding
|
0.208333 |
0.208333 |
1
|
5
|
|
GO:0005215
|
transporter activity
|
0.213704 |
0.213704 |
1
|
852
|
|
GO:0004952
|
dopamine receptor activity
|
0.217391 |
0.217391 |
1
|
5
|
|
GO:0006811
|
ion transport
|
0.221109 |
0.221109 |
1
|
331
|
|
GO:0030802
|
regulation of cyclic nucleotide biosynthetic process
|
0.222222 |
0.222222 |
1
|
8
|
|
GO:0034404
|
nucleobase, nucleoside and nucleotide biosynthetic process
|
0.223795 |
0.223795 |
2
|
163
|
|
GO:0046961
|
proton-transporting ATPase activity, rotational mechanism
|
0.224299 |
0.224299 |
1
|
24
|
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.248286 |
0.248286 |
2
|
1959
|
|
GO:0007165
|
signal transduction
|
0.254057 |
0.254057 |
1
|
548
|
|
GO:0016020
|
membrane
|
0.271576 |
0.271576 |
2
|
2122
|
|
GO:0044249
|
cellular biosynthetic process
|
0.277461 |
0.277461 |
2
|
2034
|
|
GO:0006163
|
purine nucleotide metabolic process
|
0.278592 |
0.278592 |
2
|
176
|
|
GO:0005737
|
cytoplasm
|
0.294170 |
0.294170 |
2
|
2378
|
|
GO:0010578
|
regulation of adenylate cyclase activity involved in G-protein signaling pathway
|
0.300000 |
0.300000 |
1
|
3
|
|
GO:0016817
|
hydrolase activity, acting on acid anhydrides
|
0.301217 |
0.301217 |
1
|
594
|
|
GO:0003824
|
catalytic activity
|
0.310936 |
0.310936 |
2
|
4106
|
|
GO:0007186
|
G-protein coupled receptor protein signaling pathway
|
0.318910 |
0.318910 |
1
|
199
|
|
GO:0005575
|
cellular_component
|
0.330897 |
0.330897 |
3
|
8817
|
|
GO:0007189
|
activation of adenylate cyclase activity by G-protein signaling pathway
|
0.333333 |
0.333333 |
1
|
3
|
|
GO:0023052
|
signaling
|
0.338079 |
0.338079 |
1
|
1364
|
|
GO:0005623
|
cell
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0044464
|
cell part
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0009058
|
biosynthetic process
|
0.355414 |
0.355414 |
2
|
2090
|
|
GO:0006807
|
nitrogen compound metabolic process
|
0.356248 |
0.356248 |
2
|
2093
|
|
GO:0051234
|
establishment of localization
|
0.377001 |
0.377001 |
1
|
1549
|
|
GO:0015992
|
proton transport
|
0.381818 |
0.381818 |
1
|
63
|
|
GO:0001588
|
dopamine receptor activity, coupled via Gs
|
0.400000 |
0.400000 |
1
|
2
|
|
GO:0005769
|
early endosome
|
0.400000 |
0.400000 |
1
|
14
|
|
GO:0000221
|
vacuolar proton-transporting V-type ATPase, V1 domain
|
0.400000 |
0.400000 |
1
|
16
|
|
GO:0033180
|
proton-transporting V-type ATPase, V1 domain
|
0.404255 |
0.404255 |
1
|
19
|
|
GO:0043085
|
positive regulation of catalytic activity
|
0.405063 |
0.405063 |
1
|
64
|
|
GO:0044459
|
plasma membrane part
|
0.417450 |
0.417450 |
1
|
373
|
|
GO:0080090
|
regulation of primary metabolic process
|
0.418200 |
0.418200 |
1
|
1039
|
|
GO:0009165
|
nucleotide biosynthetic process
|
0.419329 |
0.419329 |
2
|
153
|
|
GO:0031323
|
regulation of cellular metabolic process
|
0.429576 |
0.429576 |
1
|
1110
|
|
GO:0008553
|
hydrogen-exporting ATPase activity, phosphorylative mechanism
|
0.430894 |
0.430894 |
1
|
53
|
|
GO:0023034
|
intracellular signaling pathway
|
0.430962 |
0.430962 |
1
|
412
|
|
GO:0006164
|
purine nucleotide biosynthetic process
|
0.436599 |
0.436599 |
2
|
131
|
|
GO:0043492
|
ATPase activity, coupled to movement of substances
|
0.437107 |
0.437107 |
1
|
139
|
|
GO:0044093
|
positive regulation of molecular function
|
0.437126 |
0.437126 |
1
|
73
|
|
GO:0051179
|
localization
|
0.445834 |
0.445834 |
1
|
1896
|
|
GO:0015399
|
primary active transmembrane transporter activity
|
0.452778 |
0.452778 |
1
|
163
|
|
GO:0009123
|
nucleoside monophosphate metabolic process
|
0.480708 |
0.480708 |
1
|
63
|
|
GO:0019935
|
cyclic-nucleotide-mediated signaling
|
0.482759 |
0.482759 |
1
|
14
|
|
GO:0044237
|
cellular metabolic process
|
0.486483 |
0.486483 |
2
|
3814
|
|
GO:0022804
|
active transmembrane transporter activity
|
0.495868 |
0.495868 |
1
|
360
|
|
GO:0035556
|
intracellular signal transduction
|
0.499208 |
0.499208 |
1
|
315
|
|
GO:0010579
|
positive regulation of adenylate cyclase activity by G-protein signaling pathway
|
0.500000 |
0.500000 |
1
|
3
|
|
GO:0001590
|
dopamine D1 receptor activity
|
0.500000 |
0.500000 |
1
|
1
|
|
GO:0007191
|
activation of adenylate cyclase activity by dopamine receptor signaling pathway
|
0.500000 |
0.500000 |
1
|
2
|
|
GO:0022890
|
inorganic cation transmembrane transporter activity
|
0.502439 |
0.502439 |
1
|
206
|
|
GO:0019222
|
regulation of metabolic process
|
0.507779 |
0.507779 |
1
|
1242
|
|
GO:0050789
|
regulation of biological process
|
0.509927 |
0.509927 |
1
|
2246
|
|
GO:0050794
|
regulation of cellular process
|
0.518911 |
0.518911 |
1
|
2034
|
|
GO:0030814
|
regulation of cAMP metabolic process
|
0.533333 |
0.533333 |
1
|
8
|
|
GO:0007188
|
G-protein signaling, coupled to cAMP nucleotide second messenger
|
0.538462 |
0.538462 |
1
|
7
|
|
GO:0009201
|
ribonucleoside triphosphate biosynthetic process
|
0.553333 |
0.553333 |
1
|
83
|
|
GO:0032553
|
ribonucleotide binding
|
0.556624 |
0.556624 |
2
|
879
|
|
GO:0065007
|
biological regulation
|
0.561089 |
0.561089 |
1
|
2548
|
|
GO:0009206
|
purine ribonucleoside triphosphate biosynthetic process
|
0.565517 |
0.565517 |
1
|
82
|
|
GO:0032991
|
macromolecular complex
|
0.567899 |
0.567899 |
1
|
2151
|
|
GO:0009190
|
cyclic nucleotide biosynthetic process
|
0.571429 |
0.571429 |
1
|
36
|
|
GO:0030817
|
regulation of cAMP biosynthetic process
|
0.571429 |
0.571429 |
1
|
8
|
|
GO:0044422
|
organelle part
|
0.572743 |
0.572743 |
1
|
2176
|
|
GO:0032559
|
adenyl ribonucleotide binding
|
0.573921 |
0.573921 |
2
|
713
|
|
GO:0015078
|
hydrogen ion transmembrane transporter activity
|
0.574713 |
0.574713 |
1
|
100
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0033176
|
proton-transporting V-type ATPase complex
|
0.590164 |
0.590164 |
1
|
36
|
|
GO:0009889
|
regulation of biosynthetic process
|
0.599142 |
0.599142 |
1
|
888
|
|
GO:0016887
|
ATPase activity
|
0.609672 |
0.609672 |
1
|
353
|
|
GO:0004930
|
G-protein coupled receptor activity
|
0.610127 |
0.610127 |
1
|
241
|
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.617799 |
0.617799 |
1
|
905
|
|
GO:0009187
|
cyclic nucleotide metabolic process
|
0.619048 |
0.619048 |
1
|
39
|
|
GO:0009124
|
nucleoside monophosphate biosynthetic process
|
0.622829 |
0.622829 |
1
|
60
|
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.635595 |
0.635595 |
1
|
888
|
|
GO:0016405
|
CoA-ligase activity
|
0.636364 |
0.636364 |
1
|
14
|
|
GO:0016878
|
acid-thiol ligase activity
|
0.636364 |
0.636364 |
1
|
14
|
|
GO:0046034
|
ATP metabolic process
|
0.637097 |
0.637097 |
1
|
79
|
|
GO:0017076
|
purine nucleotide binding
|
0.644765 |
0.644765 |
2
|
946
|
|
GO:0007166
|
cell surface receptor linked signaling pathway
|
0.652720 |
0.652720 |
1
|
624
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0045762
|
positive regulation of adenylate cyclase activity
|
0.666667 |
0.666667 |
1
|
6
|
|
GO:0015672
|
monovalent inorganic cation transport
|
0.668016 |
0.668016 |
1
|
165
|
|
GO:0009142
|
nucleoside triphosphate biosynthetic process
|
0.668864 |
0.668864 |
1
|
83
|
|
GO:0044444
|
cytoplasmic part
|
0.669378 |
0.669378 |
1
|
1863
|
|
GO:0030554
|
adenyl nucleotide binding
|
0.670996 |
0.670996 |
2
|
775
|
|
GO:0009987
|
cellular process
|
0.673638 |
0.673638 |
2
|
6560
|
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
0.699519 |
0.699519 |
1
|
903
|
|
GO:0023033
|
signaling pathway
|
0.700880 |
0.700880 |
1
|
956
|
|
GO:0015986
|
ATP synthesis coupled proton transport
|
0.717949 |
0.717949 |
1
|
56
|
|
GO:0009145
|
purine nucleoside triphosphate biosynthetic process
|
0.721879 |
0.721879 |
1
|
82
|
|
GO:0043234
|
protein complex
|
0.727104 |
0.727104 |
1
|
1564
|
|
GO:0016787
|
hydrolase activity
|
0.729944 |
0.729944 |
1
|
1972
|
|
GO:0044446
|
intracellular organelle part
|
0.743053 |
0.743053 |
1
|
2162
|
|
GO:0006812
|
cation transport
|
0.746224 |
0.746224 |
1
|
247
|
|
GO:0016471
|
vacuolar proton-transporting V-type ATPase complex
|
0.780488 |
0.780488 |
1
|
32
|
|
GO:0043226
|
organelle
|
0.785296 |
0.785296 |
1
|
3537
|
|
GO:0022891
|
substrate-specific transmembrane transporter activity
|
0.788146 |
0.788146 |
1
|
625
|
|
GO:0044424
|
intracellular part
|
0.793631 |
0.793631 |
2
|
4384
|
|
GO:0015985
|
energy coupled proton transport, down electrochemical gradient
|
0.800000 |
0.800000 |
1
|
56
|
|
GO:0009260
|
ribonucleotide biosynthetic process
|
0.802220 |
0.802220 |
1
|
108
|
|
GO:0009141
|
nucleoside triphosphate metabolic process
|
0.805310 |
0.805310 |
1
|
126
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0008324
|
cation transmembrane transporter activity
|
0.808679 |
0.808679 |
1
|
410
|
|
GO:0015075
|
ion transmembrane transporter activity
|
0.811200 |
0.811200 |
1
|
507
|
|
GO:0022892
|
substrate-specific transporter activity
|
0.815728 |
0.815728 |
1
|
695
|
|
GO:0009152
|
purine ribonucleotide biosynthetic process
|
0.816333 |
0.816333 |
1
|
102
|
|
GO:0004872
|
receptor activity
|
0.821109 |
0.821109 |
1
|
459
|
|
GO:0006753
|
nucleoside phosphate metabolic process
|
0.823455 |
0.823455 |
2
|
226
|
|
GO:0009199
|
ribonucleoside triphosphate metabolic process
|
0.827815 |
0.827815 |
1
|
125
|
|
GO:0007190
|
activation of adenylate cyclase activity
|
0.833333 |
0.833333 |
1
|
5
|
|
GO:0015077
|
monovalent inorganic cation transmembrane transporter activity
|
0.844660 |
0.844660 |
1
|
174
|
|
GO:0022857
|
transmembrane transporter activity
|
0.852113 |
0.852113 |
1
|
726
|
|
GO:0042626
|
ATPase activity, coupled to transmembrane movement of substances
|
0.852761 |
0.852761 |
1
|
139
|
|
GO:0009205
|
purine ribonucleoside triphosphate metabolic process
|
0.855172 |
0.855172 |
1
|
124
|
|
GO:0005622
|
intracellular
|
0.859425 |
0.859425 |
2
|
4734
|
|
GO:0004888
|
transmembrane receptor activity
|
0.860566 |
0.860566 |
1
|
395
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.863297 |
0.863297 |
2
|
879
|
|
GO:0009259
|
ribonucleotide metabolic process
|
0.887906 |
0.887906 |
1
|
150
|
|
GO:0045761
|
regulation of adenylate cyclase activity
|
0.888889 |
0.888889 |
1
|
8
|
|
GO:0042623
|
ATPase activity, coupled
|
0.900850 |
0.900850 |
1
|
318
|
|
GO:0009144
|
purine nucleoside triphosphate metabolic process
|
0.909160 |
0.909160 |
1
|
124
|
|
GO:0044238
|
primary metabolic process
|
0.914505 |
0.914505 |
2
|
4259
|
|
GO:0015662
|
ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism
|
0.916667 |
0.916667 |
1
|
77
|
|
GO:0031224
|
intrinsic to membrane
|
0.924695 |
0.924695 |
1
|
1014
|
|
GO:0019933
|
cAMP-mediated signaling
|
0.928571 |
0.928571 |
1
|
13
|
|
GO:0006754
|
ATP biosynthetic process
|
0.939759 |
0.939759 |
1
|
78
|
|
GO:0009150
|
purine ribonucleotide metabolic process
|
0.957319 |
0.957319 |
1
|
144
|
|
GO:0043229
|
intracellular organelle
|
0.961659 |
0.961659 |
1
|
3530
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0017111
|
nucleoside-triphosphatase activity
|
0.983022 |
0.983022 |
1
|
579
|
|
GO:0016021
|
integral to membrane
|
0.984221 |
0.984221 |
1
|
998
|
|
GO:0001883
|
purine nucleoside binding
|
0.984743 |
0.984743 |
2
|
778
|
|
GO:0016462
|
pyrophosphatase activity
|
0.991582 |
0.991582 |
1
|
589
|
|
GO:0005524
|
ATP binding
|
1.00000 |
1.00000 |
1
|
712
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|
|
GO:0016818
|
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
|
1.00000 |
1.00000 |
1
|
594
|
|
GO:0004871
|
signal transducer activity
|
1.00000 |
1.00000 |
1
|
559
|
|
GO:0023060
|
signal transmission
|
1.00000 |
1.00000 |
1
|
760
|
|
GO:0009117
|
nucleotide metabolic process
|
1.00000 |
1.00000 |
2
|
226
|
|
GO:0015405
|
P-P-bond-hydrolysis-driven transmembrane transporter activity
|
1.00000 |
1.00000 |
1
|
163
|