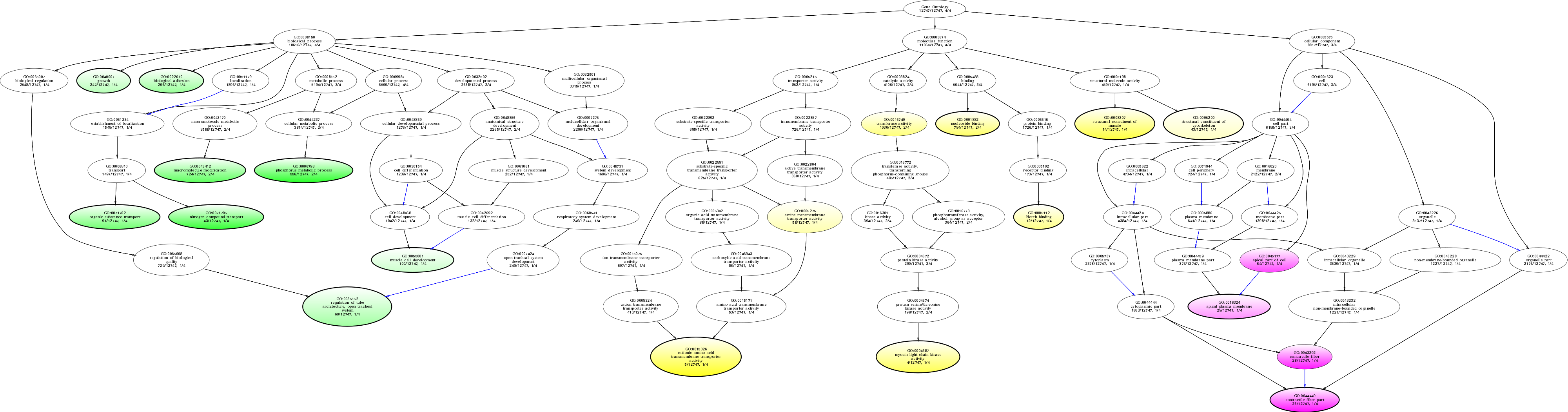
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0044449
|
contractile fiber part
|
0.00848010 |
0.00848010 |
1
|
26
|
|
GO:0043292
|
contractile fiber
|
0.0107157 |
0.0107157 |
1
|
28
|
|
GO:0015326
|
cationic amino acid transmembrane transporter activity
|
0.0115741 |
0.0115741 |
1
|
5
|
|
GO:0006793
|
phosphorus metabolic process
|
0.0212188 |
0.0212188 |
2
|
556
|
|
GO:0071705
|
nitrogen compound transport
|
0.0287241 |
0.0287241 |
1
|
43
|
|
GO:0008307
|
structural constituent of muscle
|
0.0305011 |
0.0305011 |
1
|
14
|
|
GO:0045177
|
apical part of cell
|
0.0306786 |
0.0306786 |
1
|
64
|
|
GO:0004687
|
myosin light chain kinase activity
|
0.0398965 |
0.0398965 |
1
|
4
|
|
GO:0043412
|
macromolecule modification
|
0.0406718 |
0.0406718 |
2
|
724
|
|
GO:0001882
|
nucleoside binding
|
0.0525332 |
0.0525332 |
2
|
784
|
|
GO:0071702
|
organic substance transport
|
0.0607882 |
0.0607882 |
1
|
91
|
|
GO:0016740
|
transferase activity
|
0.0628812 |
0.0628812 |
2
|
1030
|
|
GO:0005112
|
Notch binding
|
0.0693642 |
0.0693642 |
1
|
12
|
|
GO:0016324
|
apical plasma membrane
|
0.0719603 |
0.0719603 |
1
|
29
|
|
GO:0022610
|
biological adhesion
|
0.0753989 |
0.0753989 |
1
|
206
|
|
GO:0035152
|
regulation of tube architecture, open tracheal system
|
0.0782313 |
0.0782313 |
1
|
69
|
|
GO:0005275
|
amine transmembrane transporter activity
|
0.0845481 |
0.0845481 |
1
|
58
|
|
GO:0040007
|
growth
|
0.0898814 |
0.0898814 |
1
|
247
|
|
GO:0005200
|
structural constituent of cytoskeleton
|
0.0936819 |
0.0936819 |
1
|
43
|
|
GO:0055001
|
muscle cell development
|
0.0938967 |
0.0938967 |
1
|
100
|
|
GO:0005102
|
receptor binding
|
0.100232 |
0.100232 |
1
|
173
|
|
GO:0042692
|
muscle cell differentiation
|
0.102644 |
0.102644 |
1
|
132
|
|
GO:0030029
|
actin filament-based process
|
0.108705 |
0.108705 |
1
|
186
|
|
GO:0007155
|
cell adhesion
|
0.111846 |
0.111846 |
1
|
192
|
|
GO:0006468
|
protein phosphorylation
|
0.112462 |
0.112462 |
2
|
277
|
|
GO:0000166
|
nucleotide binding
|
0.112563 |
0.112563 |
2
|
1178
|
|
GO:0030239
|
myofibril assembly
|
0.117886 |
0.117886 |
1
|
29
|
|
GO:0010927
|
cellular component assembly involved in morphogenesis
|
0.134157 |
0.134157 |
1
|
152
|
|
GO:0005342
|
organic acid transmembrane transporter activity
|
0.140800 |
0.140800 |
1
|
88
|
|
GO:0005509
|
calcium ion binding
|
0.144237 |
0.144237 |
1
|
209
|
|
GO:0009987
|
cellular process
|
0.145754 |
0.145754 |
4
|
6560
|
|
GO:0060541
|
respiratory system development
|
0.146816 |
0.146816 |
1
|
249
|
|
GO:0005198
|
structural molecule activity
|
0.155920 |
0.155920 |
1
|
459
|
|
GO:0007498
|
mesoderm development
|
0.170557 |
0.170557 |
1
|
95
|
|
GO:0060089
|
molecular transducer activity
|
0.187314 |
0.187314 |
1
|
559
|
|
GO:0006629
|
lipid metabolic process
|
0.191945 |
0.191945 |
1
|
292
|
|
GO:0032989
|
cellular component morphogenesis
|
0.193108 |
0.193108 |
1
|
566
|
|
GO:0044267
|
cellular protein metabolic process
|
0.193579 |
0.193579 |
2
|
1505
|
|
GO:0006464
|
protein modification process
|
0.196094 |
0.196094 |
2
|
684
|
|
GO:0061061
|
muscle structure development
|
0.210182 |
0.210182 |
1
|
252
|
|
GO:0016772
|
transferase activity, transferring phosphorus-containing groups
|
0.230717 |
0.230717 |
2
|
495
|
|
GO:0044459
|
plasma membrane part
|
0.236675 |
0.236675 |
1
|
373
|
|
GO:0055085
|
transmembrane transport
|
0.246988 |
0.246988 |
1
|
469
|
|
GO:0032502
|
developmental process
|
0.259158 |
0.259158 |
2
|
2638
|
|
GO:0016020
|
membrane
|
0.271576 |
0.271576 |
2
|
2122
|
|
GO:0005215
|
transporter activity
|
0.274270 |
0.274270 |
1
|
852
|
|
GO:0022607
|
cellular component assembly
|
0.275418 |
0.275418 |
1
|
577
|
|
GO:0065008
|
regulation of biological quality
|
0.286107 |
0.286107 |
1
|
729
|
|
GO:0008152
|
metabolic process
|
0.296531 |
0.296531 |
3
|
5194
|
|
GO:0044085
|
cellular component biogenesis
|
0.299810 |
0.299810 |
1
|
632
|
|
GO:0031032
|
actomyosin structure organization
|
0.306011 |
0.306011 |
1
|
56
|
|
GO:0071944
|
cell periphery
|
0.311271 |
0.311271 |
1
|
724
|
|
GO:0048646
|
anatomical structure formation involved in morphogenesis
|
0.322148 |
0.322148 |
1
|
466
|
|
GO:0005488
|
binding
|
0.327389 |
0.327389 |
3
|
5641
|
|
GO:0030036
|
actin cytoskeleton organization
|
0.339518 |
0.339518 |
1
|
183
|
|
GO:0005623
|
cell
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0044464
|
cell part
|
0.346816 |
0.346816 |
3
|
6195
|
|
GO:0043228
|
non-membrane-bounded organelle
|
0.346904 |
0.346904 |
1
|
1227
|
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.347592 |
0.347592 |
1
|
1227
|
|
GO:0045214
|
sarcomere organization
|
0.357143 |
0.357143 |
1
|
20
|
|
GO:0071844
|
cellular component assembly at cellular level
|
0.373193 |
0.373193 |
1
|
568
|
|
GO:0044260
|
cellular macromolecule metabolic process
|
0.408435 |
0.408435 |
2
|
2878
|
|
GO:0044444
|
cytoplasmic part
|
0.424954 |
0.424954 |
1
|
1863
|
|
GO:0009888
|
tissue development
|
0.431439 |
0.431439 |
1
|
557
|
|
GO:0007010
|
cytoskeleton organization
|
0.453469 |
0.453469 |
1
|
536
|
|
GO:0019538
|
protein metabolic process
|
0.463198 |
0.463198 |
2
|
2053
|
|
GO:0051234
|
establishment of localization
|
0.467930 |
0.467930 |
1
|
1549
|
|
GO:0004674
|
protein serine/threonine kinase activity
|
0.470135 |
0.470135 |
2
|
199
|
|
GO:0015837
|
amine transport
|
0.472527 |
0.472527 |
1
|
43
|
|
GO:0003824
|
catalytic activity
|
0.474372 |
0.474372 |
2
|
4106
|
|
GO:0008150
|
biological_process
|
0.481028 |
0.481028 |
4
|
10616
|
|
GO:0022804
|
active transmembrane transporter activity
|
0.495868 |
0.495868 |
1
|
360
|
|
GO:0005886
|
plasma membrane
|
0.498869 |
0.498869 |
1
|
641
|
|
GO:0004672
|
protein kinase activity
|
0.530423 |
0.530423 |
2
|
290
|
|
GO:0044425
|
membrane part
|
0.535780 |
0.535780 |
1
|
1398
|
|
GO:0030018
|
Z disc
|
0.538462 |
0.538462 |
1
|
14
|
|
GO:0031674
|
I band
|
0.538462 |
0.538462 |
1
|
14
|
|
GO:0016773
|
phosphotransferase activity, alcohol group as acceptor
|
0.540351 |
0.540351 |
2
|
364
|
|
GO:0048869
|
cellular developmental process
|
0.540817 |
0.540817 |
1
|
1276
|
|
GO:0005737
|
cytoplasm
|
0.542427 |
0.542427 |
1
|
2378
|
|
GO:0051179
|
localization
|
0.544835 |
0.544835 |
1
|
1896
|
|
GO:0044238
|
primary metabolic process
|
0.551267 |
0.551267 |
3
|
4259
|
|
GO:0032553
|
ribonucleotide binding
|
0.556624 |
0.556624 |
2
|
879
|
|
GO:0003674
|
molecular_function
|
0.567526 |
0.567526 |
4
|
11064
|
|
GO:0015849
|
organic acid transport
|
0.571429 |
0.571429 |
1
|
52
|
|
GO:0044422
|
organelle part
|
0.572743 |
0.572743 |
1
|
2176
|
|
GO:0032559
|
adenyl ribonucleotide binding
|
0.573921 |
0.573921 |
2
|
713
|
|
GO:0016310
|
phosphorylation
|
0.575740 |
0.575740 |
2
|
422
|
|
GO:0015171
|
amino acid transmembrane transporter activity
|
0.582418 |
0.582418 |
1
|
53
|
|
GO:0071840
|
cellular component organization or biogenesis
|
0.587518 |
0.587518 |
1
|
2108
|
|
GO:0043167
|
ion binding
|
0.603911 |
0.603911 |
1
|
1498
|
|
GO:0016301
|
kinase activity
|
0.633223 |
0.633223 |
2
|
394
|
|
GO:0005575
|
cellular_component
|
0.637024 |
0.637024 |
3
|
8817
|
|
GO:0017076
|
purine nucleotide binding
|
0.644765 |
0.644765 |
2
|
946
|
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
0.661861 |
0.661861 |
1
|
1562
|
|
GO:0005515
|
protein binding
|
0.665786 |
0.665786 |
1
|
1726
|
|
GO:0065007
|
biological regulation
|
0.666464 |
0.666464 |
1
|
2548
|
|
GO:0030554
|
adenyl nucleotide binding
|
0.670996 |
0.670996 |
2
|
775
|
|
GO:0044237
|
cellular metabolic process
|
0.673909 |
0.673909 |
2
|
3814
|
|
GO:0048468
|
cell development
|
0.705715 |
0.705715 |
1
|
1042
|
|
GO:0006865
|
amino acid transport
|
0.709091 |
0.709091 |
1
|
39
|
|
GO:0071842
|
cellular component organization at cellular level
|
0.724105 |
0.724105 |
1
|
1517
|
|
GO:0048856
|
anatomical structure development
|
0.737157 |
0.737157 |
2
|
2265
|
|
GO:0043170
|
macromolecule metabolic process
|
0.772351 |
0.772351 |
2
|
3588
|
|
GO:0032501
|
multicellular organismal process
|
0.776347 |
0.776347 |
1
|
3315
|
|
GO:0006996
|
organelle organization
|
0.779169 |
0.779169 |
1
|
1182
|
|
GO:0043226
|
organelle
|
0.785296 |
0.785296 |
1
|
3537
|
|
GO:0022891
|
substrate-specific transmembrane transporter activity
|
0.788146 |
0.788146 |
1
|
625
|
|
GO:0055002
|
striated muscle cell development
|
0.798387 |
0.798387 |
1
|
99
|
|
GO:0043229
|
intracellular organelle
|
0.804100 |
0.804100 |
1
|
3530
|
|
GO:0008324
|
cation transmembrane transporter activity
|
0.808679 |
0.808679 |
1
|
410
|
|
GO:0015075
|
ion transmembrane transporter activity
|
0.811200 |
0.811200 |
1
|
507
|
|
GO:0022892
|
substrate-specific transporter activity
|
0.815728 |
0.815728 |
1
|
695
|
|
GO:0004872
|
receptor activity
|
0.821109 |
0.821109 |
1
|
459
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.824951 |
0.824951 |
1
|
1534
|
|
GO:0022857
|
transmembrane transporter activity
|
0.852113 |
0.852113 |
1
|
726
|
|
GO:0030016
|
myofibril
|
0.857143 |
0.857143 |
1
|
24
|
|
GO:0032555
|
purine ribonucleotide binding
|
0.863297 |
0.863297 |
2
|
879
|
|
GO:0048731
|
system development
|
0.882863 |
0.882863 |
1
|
1696
|
|
GO:0030017
|
sarcomere
|
0.884615 |
0.884615 |
1
|
23
|
|
GO:0007275
|
multicellular organismal development
|
0.887792 |
0.887792 |
1
|
2296
|
|
GO:0051146
|
striated muscle cell differentiation
|
0.939394 |
0.939394 |
1
|
124
|
|
GO:0006810
|
transport
|
0.966430 |
0.966430 |
1
|
1497
|
|
GO:0046872
|
metal ion binding
|
0.967936 |
0.967936 |
1
|
1449
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0016043
|
cellular component organization
|
0.973435 |
0.973435 |
1
|
2052
|
|
GO:0044424
|
intracellular part
|
0.975047 |
0.975047 |
1
|
4384
|
|
GO:0046943
|
carboxylic acid transmembrane transporter activity
|
0.977273 |
0.977273 |
1
|
86
|
|
GO:0001883
|
purine nucleoside binding
|
0.984743 |
0.984743 |
2
|
778
|
|
GO:0005622
|
intracellular
|
0.986904 |
0.986904 |
1
|
4734
|
|
GO:0007424
|
open tracheal system development
|
0.995984 |
0.995984 |
1
|
248
|
|
GO:0005524
|
ATP binding
|
0.997195 |
0.997195 |
2
|
712
|
|
GO:0043169
|
cation binding
|
0.999332 |
0.999332 |
1
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
4
|
12747
|
|
GO:0004871
|
signal transducer activity
|
1.00000 |
1.00000 |
1
|
559
|
|
GO:0006796
|
phosphate metabolic process
|
1.00000 |
1.00000 |
2
|
556
|
|
GO:0046942
|
carboxylic acid transport
|
1.00000 |
1.00000 |
1
|
52
|