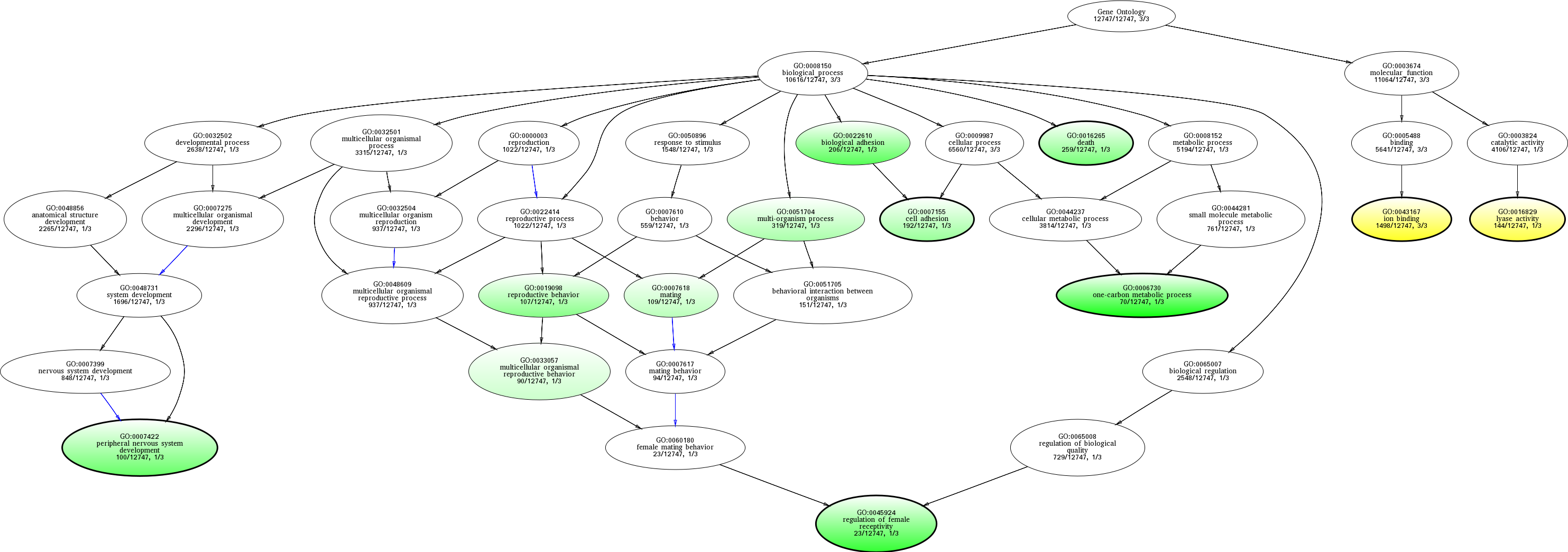
| ID | Name | p-Value | p-Value (Adj) | Study Count | Population Count |
|---|
|
GO:0006730
|
one-carbon metabolic process
|
0.0182244 |
0.0182244 |
1
|
70
|
|
GO:0043167
|
ion binding
|
0.0186994 |
0.0186994 |
3
|
1498
|
|
GO:0045924
|
regulation of female receptivity
|
0.0315501 |
0.0315501 |
1
|
23
|
|
GO:0016829
|
lyase activity
|
0.0350706 |
0.0350706 |
1
|
144
|
|
GO:0022610
|
biological adhesion
|
0.0570970 |
0.0570970 |
1
|
206
|
|
GO:0007422
|
peripheral nervous system development
|
0.0589623 |
0.0589623 |
1
|
100
|
|
GO:0016265
|
death
|
0.0714268 |
0.0714268 |
1
|
259
|
|
GO:0019098
|
reproductive behavior
|
0.0752461 |
0.0752461 |
1
|
107
|
|
GO:0007155
|
cell adhesion
|
0.0851090 |
0.0851090 |
1
|
192
|
|
GO:0051704
|
multi-organism process
|
0.0874732 |
0.0874732 |
1
|
319
|
|
GO:0007618
|
mating
|
0.0912134 |
0.0912134 |
1
|
109
|
|
GO:0033057
|
multicellular organismal reproductive behavior
|
0.0947368 |
0.0947368 |
1
|
90
|
|
GO:0061061
|
muscle structure development
|
0.111258 |
0.111258 |
1
|
252
|
|
GO:0008219
|
cell death
|
0.112158 |
0.112158 |
1
|
255
|
|
GO:0005488
|
binding
|
0.132501 |
0.132501 |
3
|
5641
|
|
GO:0044281
|
small molecule metabolic process
|
0.146515 |
0.146515 |
1
|
761
|
|
GO:0007517
|
muscle organ development
|
0.149102 |
0.149102 |
1
|
191
|
|
GO:0007525
|
somatic muscle development
|
0.166667 |
0.166667 |
1
|
42
|
|
GO:0031226
|
intrinsic to plasma membrane
|
0.179931 |
0.179931 |
1
|
208
|
|
GO:0005887
|
integral to plasma membrane
|
0.203593 |
0.203593 |
1
|
204
|
|
GO:0051705
|
behavioral interaction between organisms
|
0.209431 |
0.209431 |
1
|
151
|
|
GO:0060180
|
female mating behavior
|
0.219048 |
0.219048 |
1
|
23
|
|
GO:0071944
|
cell periphery
|
0.220095 |
0.220095 |
1
|
724
|
|
GO:0009987
|
cellular process
|
0.235914 |
0.235914 |
3
|
6560
|
|
GO:0044459
|
plasma membrane part
|
0.236675 |
0.236675 |
1
|
373
|
|
GO:0009790
|
embryo development
|
0.242987 |
0.242987 |
1
|
641
|
|
GO:0000003
|
reproduction
|
0.261920 |
0.261920 |
1
|
1022
|
|
GO:0022414
|
reproductive process
|
0.261920 |
0.261920 |
1
|
1022
|
|
GO:0048609
|
multicellular organismal reproductive process
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0032504
|
multicellular organism reproduction
|
0.279118 |
0.279118 |
1
|
937
|
|
GO:0016339
|
calcium-dependent cell-cell adhesion
|
0.283784 |
0.283784 |
1
|
21
|
|
GO:0065008
|
regulation of biological quality
|
0.286107 |
0.286107 |
1
|
729
|
|
GO:0005886
|
plasma membrane
|
0.292027 |
0.292027 |
1
|
641
|
|
GO:0046530
|
photoreceptor cell differentiation
|
0.295620 |
0.295620 |
1
|
162
|
|
GO:0001751
|
compound eye photoreceptor cell differentiation
|
0.354497 |
0.354497 |
1
|
134
|
|
GO:0007610
|
behavior
|
0.361111 |
0.361111 |
1
|
559
|
|
GO:0001754
|
eye photoreceptor cell differentiation
|
0.361386 |
0.361386 |
1
|
146
|
|
GO:0005509
|
calcium ion binding
|
0.373519 |
0.373519 |
1
|
209
|
|
GO:0009887
|
organ morphogenesis
|
0.374384 |
0.374384 |
1
|
684
|
|
GO:0016835
|
carbon-oxygen lyase activity
|
0.375000 |
0.375000 |
1
|
54
|
|
GO:0050896
|
response to stimulus
|
0.376795 |
0.376795 |
1
|
1548
|
|
GO:0007156
|
homophilic cell adhesion
|
0.378378 |
0.378378 |
1
|
28
|
|
GO:0016337
|
cell-cell adhesion
|
0.385417 |
0.385417 |
1
|
74
|
|
GO:0044425
|
membrane part
|
0.400435 |
0.400435 |
1
|
1398
|
|
GO:0004089
|
carbonate dehydratase activity
|
0.434783 |
0.434783 |
1
|
20
|
|
GO:0007423
|
sensory organ development
|
0.438003 |
0.438003 |
1
|
544
|
|
GO:0048869
|
cellular developmental process
|
0.442161 |
0.442161 |
1
|
1276
|
|
GO:0030182
|
neuron differentiation
|
0.442292 |
0.442292 |
1
|
548
|
|
GO:0003677
|
DNA binding
|
0.444205 |
0.444205 |
1
|
824
|
|
GO:0022008
|
neurogenesis
|
0.452398 |
0.452398 |
1
|
632
|
|
GO:0005623
|
cell
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0044464
|
cell part
|
0.493651 |
0.493651 |
2
|
6195
|
|
GO:0007399
|
nervous system development
|
0.500000 |
0.500000 |
1
|
848
|
|
GO:0007617
|
mating behavior
|
0.534091 |
0.534091 |
1
|
94
|
|
GO:0048592
|
eye morphogenesis
|
0.542069 |
0.542069 |
1
|
393
|
|
GO:0048513
|
organ development
|
0.548344 |
0.548344 |
1
|
1242
|
|
GO:0065007
|
biological regulation
|
0.561089 |
0.561089 |
1
|
2548
|
|
GO:0016020
|
membrane
|
0.567775 |
0.567775 |
1
|
2122
|
|
GO:0032502
|
developmental process
|
0.575616 |
0.575616 |
1
|
2638
|
|
GO:0008150
|
biological_process
|
0.577615 |
0.577615 |
3
|
10616
|
|
GO:0009653
|
anatomical structure morphogenesis
|
0.581501 |
0.581501 |
1
|
1534
|
|
GO:0008270
|
zinc ion binding
|
0.610500 |
0.610500 |
2
|
901
|
|
GO:0006915
|
apoptosis
|
0.633333 |
0.633333 |
1
|
152
|
|
GO:0005634
|
nucleus
|
0.639356 |
0.639356 |
1
|
1826
|
|
GO:0043226
|
organelle
|
0.641414 |
0.641414 |
1
|
3537
|
|
GO:0003674
|
molecular_function
|
0.653878 |
0.653878 |
3
|
11064
|
|
GO:0048731
|
system development
|
0.657619 |
0.657619 |
1
|
1696
|
|
GO:0007275
|
multicellular organismal development
|
0.664929 |
0.664929 |
1
|
2296
|
|
GO:0032501
|
multicellular organismal process
|
0.674757 |
0.674757 |
1
|
3315
|
|
GO:0003676
|
nucleic acid binding
|
0.697754 |
0.697754 |
1
|
1855
|
|
GO:0031224
|
intrinsic to membrane
|
0.725322 |
0.725322 |
1
|
1014
|
|
GO:0003824
|
catalytic activity
|
0.751316 |
0.751316 |
1
|
4106
|
|
GO:0005575
|
cellular_component
|
0.773470 |
0.773470 |
2
|
8817
|
|
GO:0043229
|
intracellular organelle
|
0.804100 |
0.804100 |
1
|
3530
|
|
GO:0043227
|
membrane-bounded organelle
|
0.808312 |
0.808312 |
1
|
2859
|
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.808607 |
0.808607 |
1
|
2856
|
|
GO:0001745
|
compound eye morphogenesis
|
0.811947 |
0.811947 |
1
|
367
|
|
GO:0001654
|
eye development
|
0.841912 |
0.841912 |
1
|
458
|
|
GO:0016836
|
hydro-lyase activity
|
0.851852 |
0.851852 |
1
|
46
|
|
GO:0048856
|
anatomical structure development
|
0.858605 |
0.858605 |
1
|
2265
|
|
GO:0008152
|
metabolic process
|
0.866808 |
0.866808 |
1
|
5194
|
|
GO:0044237
|
cellular metabolic process
|
0.868168 |
0.868168 |
1
|
3814
|
|
GO:0046914
|
transition metal ion binding
|
0.892059 |
0.892059 |
2
|
1153
|
|
GO:0046872
|
metal ion binding
|
0.906799 |
0.906799 |
3
|
1449
|
|
GO:0044424
|
intracellular part
|
0.914575 |
0.914575 |
1
|
4384
|
|
GO:0048749
|
compound eye development
|
0.932314 |
0.932314 |
1
|
427
|
|
GO:0012501
|
programmed cell death
|
0.941176 |
0.941176 |
1
|
240
|
|
GO:0005622
|
intracellular
|
0.944411 |
0.944411 |
1
|
4734
|
|
GO:0048699
|
generation of neurons
|
0.954114 |
0.954114 |
1
|
603
|
|
GO:0030154
|
cell differentiation
|
0.971003 |
0.971003 |
1
|
1239
|
|
GO:0016021
|
integral to membrane
|
0.984221 |
0.984221 |
1
|
998
|
|
GO:0043169
|
cation binding
|
0.997997 |
0.997997 |
3
|
1497
|
|
GO:0000000
|
root
|
1.00000 |
1.00000 |
3
|
12747
|